For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTKPTDAPTRTEDLVEIQQLLAKYAVTITQGDIDGLISVFTPDGTYSAFGETYTLNRFPVLVDAAPKGLF MTGTALVDLVQGADTATGTQPLCFIEHSTHDMRIGYYRDTYERTADGWRLKTRSMTFIRRNGDHDHGRPH AIGRPEA
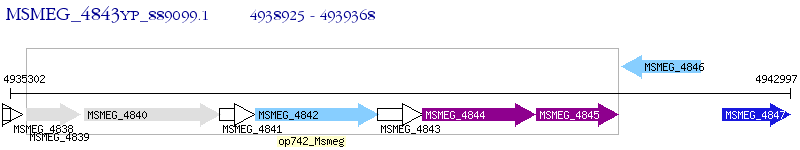
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4843 | - | - | 100% (147) | hypothetical protein MSMEG_4843 |
| M. smegmatis MC2 155 | MSMEG_2434 | - | 3e-05 | 31.30% (131) | hypothetical protein MSMEG_2434 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2934c | - | 1e-05 | 32.33% (133) | hypothetical protein Mb2934c |
| M. gilvum PYR-GCK | Mflv_2436 | - | 1e-64 | 83.45% (139) | hypothetical protein Mflv_2436 |
| M. tuberculosis H37Rv | Rv2910c | - | 1e-05 | 32.33% (133) | hypothetical protein Rv2910c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4747 | - | 3e-62 | 78.99% (138) | hypothetical protein MMAR_4747 |
| M. avium 104 | MAV_0902 | - | 2e-63 | 82.48% (137) | hypothetical protein MAV_0902 |
| M. thermoresistible (build 8) | TH_2021 | - | 1e-63 | 82.86% (140) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2046 | - | 5e-07 | 32.31% (130) | hypothetical protein MUL_2046 |
| M. vanbaalenii PYR-1 | Mvan_4216 | - | 2e-70 | 88.57% (140) | hypothetical protein Mvan_4216 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4843|M.smegmatis_MC2_155 ----MTKPTDAPTRTEDLVEIQQLLAKYAVTITQGDIDGLISVFTPDG--
Mvan_4216|M.vanbaalenii_PYR-1 ----MST---QPTRTEDLVEIQQLLAKYAVTITQGDIDGLISVFTPDG--
Mflv_2436|M.gilvum_PYR-GCK ----MTS------RTEDLVEIQQLLAKYAVTITQGDLDGLISVFTPDG--
TH_2021|M.thermoresistible__bu ----MT-------RTDDLVEIQQLLAKYAVTITQGDIDGLISVFTPDG--
MMAR_4747|M.marinum_M ----MS------TKTDDLVEIQQLLARYAVTITQNDIEGLVAVFTPDG--
MAV_0902|M.avium_104 ----MST-----TRTDDLVEIQQLLARYAVTITQEDIDGLVAVFTPDG--
Mb2934c|M.bovis_AF2122/97 MCAVLDRSMLSVAEISDRLEIQQLLVDYSSAIDQRRFDDLDRVFTPDAYI
Rv2910c|M.tuberculosis_H37Rv MCAVLDRSMLSVAEISDRLEIQQLLVDYSSAIDQRRFDDLDRVFTPDAYI
MUL_2046|M.ulcerans_Agy99 ---MLNRIMLSLPEISDRLEIQQLLVDYSTAIDLRRFDDLDKVFTPDAYI
: . .* :******. *: :* ::.* *****.
MSMEG_4843|M.smegmatis_MC2_155 TYSAFG-----ETYTLNRFPVLVDAAPKGLFMTGTALVDLVQGA--DTAT
Mvan_4216|M.vanbaalenii_PYR-1 TYSAFG-----STYTLARFPELVDAAPKGLFMTGTALVDLNEGE--DTAS
Mflv_2436|M.gilvum_PYR-GCK TYSAFG-----STYTLARFPELVDAAPKGLFMTGTSLVTLDADDP-DKAT
TH_2021|M.thermoresistible__bu SYSAFG-----STYTLARFPELVAAAPKGLFMTGESLVTFDADDP-DKAS
MMAR_4747|M.marinum_M TYSAFG-----DTYGLDRFPDLVAAAPKGLFLTATAVVELAGDS----AT
MAV_0902|M.avium_104 TYSAFG-----ETYSLNRFPELVAAAPKGLFLTGTAVVDLDGDQ----AS
Mb2934c|M.bovis_AF2122/97 DYRALGGIDGRYPKIKQWLSQVLGNFPVYAHMLGNFSVRVDGDTASSRVI
Rv2910c|M.tuberculosis_H37Rv DYRALGGIDGRYPKIKQWLSQVLGNFPVYAHMLGNFSVRVDGDTASSRVI
MUL_2046|M.ulcerans_Agy99 DYRAMGGIDGQYPHVKAWLAEVLPNFANYAHMLGLPSIRLAGDTATARTF
* *:* . :. :: . .: . : . . .
MSMEG_4843|M.smegmatis_MC2_155 GTQPLCFIEHSTHDMRIG-YYRDTYERTADGWRLKTRSMTFIRRNGDHDH
Mvan_4216|M.vanbaalenii_PYR-1 GTQPLCFIEHSKHDMRIG-YYRDTYVRSDDGWRLRTRAMTFIRRSGEHDS
Mflv_2436|M.gilvum_PYR-GCK GTQPLCFIEHSKHDMRIG-YYNDTYVRTEDGWRLRTRAMTFIRRSGEHDS
TH_2021|M.thermoresistible__bu GTQPLCFIEHSTHDMRIG-YYNDTYVRTADGWRLRTRAMTFIRRSGAHDS
MMAR_4747|M.marinum_M GTQPLCFIEHATHDMRIG-YYRDTYARTADGWRLKTRAMTFIRRSGVHDS
MAV_0902|M.avium_104 GTQPLCFIDHATHDMRIG-YYRDTYVRTPDGWRLKTRAMTFIRRSGVHDS
Mb2934c|M.bovis_AF2122/97 CFNPMVFAGDRQQVLFCGLWYDDDFVRTPDGWRIIRRVETKCFQKMM---
Rv2910c|M.tuberculosis_H37Rv CFNPMVFAGDRQQVLFCGLWYDDDFVRTPDGWRIIRRVETKCFQKMM---
MUL_2046|M.ulcerans_Agy99 CFNPMVFAGEKPTTMLLGLWYDDEFVRTAEGWRMSRRAETKCFDRRP---
:*: * . : * :* * : *: :***: * *
MSMEG_4843|M.smegmatis_MC2_155 GRPHAIGRPE----A-------
Mvan_4216|M.vanbaalenii_PYR-1 GRPHAIGRPE----AG------
Mflv_2436|M.gilvum_PYR-GCK GRPHAIGRPDGDVAAGAAPGAR
TH_2021|M.thermoresistible__bu GRPHAIGRPE----AG------
MMAR_4747|M.marinum_M GRPHAVGRPQP-----------
MAV_0902|M.avium_104 GRPHAIGRPAGDSAAQAAT---
Mb2934c|M.bovis_AF2122/97 ----------------------
Rv2910c|M.tuberculosis_H37Rv ----------------------
MUL_2046|M.ulcerans_Agy99 ----------------------