For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSRILVGSAPDSWGVWFPDDPQQTPYTRFLDEVASAGYRWIELGPYGYLPTDPDKLADELAQRDLSLSAG TVFEHLHQSATRGDDAWNSVWTQIEDVARLTAAVGGKHVVVIPEMWRDPATGAVLEDRNLTAEQWRQKTQ GMNELGRAMFEKYGVRAQYHPHADSHVDTEENVYRFLDGTDGEFVNLCLDTGHISYCGGDNIAIIRHAPE RIGYLHLKQVDPEVRAKVEAEDLPFGEAVKLGAMTEPPRGIPDMPPLLAEIERLGVDVFAIVEQDMYPCD PGSPLPIAQRTRSYLRSCGVPAVRFT
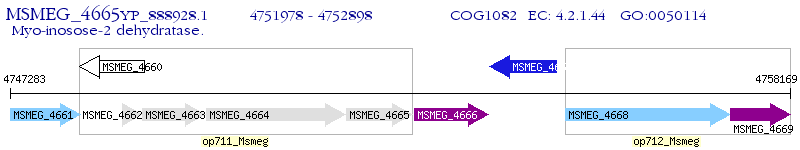
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4665 | - | - | 100% (306) | IolE protein |
| M. smegmatis MC2 155 | MSMEG_4651 | - | 1e-27 | 33.00% (297) | AP endonuclease, family protein 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2612 | - | 3e-25 | 31.69% (284) | xylose isomerase domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0386 | - | 1e-163 | 86.89% (305) | xylose isomerase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4665|M.smegmatis_MC2_155 MSRILVGSAPDSWGVWFPDDP-QQTPYTRFLDEVASAGYRWIELGPYGYL
Mvan_0386|M.vanbaalenii_PYR-1 MSTIRVGSAPDSWGVWFPDDP-QQTPYSRFLDEVAESGYEWIELGPYGYL
Mflv_2612|M.gilvum_PYR-GCK ---MKIAGAPISWGVCEVPGWGHQLSADRVLTEMRQAGLTATELGPEGFL
: :..** **** . :* . *.* *: .:* **** *:*
MSMEG_4665|M.smegmatis_MC2_155 PTDPDKLADELAQRDLSLSAGTVFEHLHQSATRGDDAWNSVWTQIEDVAR
Mvan_0386|M.vanbaalenii_PYR-1 PTDPAKLSDELAARGLKLSAGTVFEHLHQSGSRGDDAWNSVWRQIEDVAK
Mflv_2612|M.gilvum_PYR-GCK PADPAELVKLLDGYGLACVGGFVPVVLFSDDHDPADG-------LAGPLE
*:** :* . * .* .* * *... *. : . .
MSMEG_4665|M.smegmatis_MC2_155 LTAAVGGKHVVVIPEMWRDPATGAVLEDRNLTAEQWRQKTQGMNELGRAM
Mvan_0386|M.vanbaalenii_PYR-1 LTAAVGGTHVVVIPEMWRDPATGEVLEDRNLTAEQWAKKTAGMNDLGKAM
Mflv_2612|M.gilvum_PYR-GCK SLVAAGAEVVVLAAATGQDGYD----RRPVLDEKQWATLLSNLDRLADLV
.*.*. **: . :* . * :** .:: *. :
MSMEG_4665|M.smegmatis_MC2_155 FEKYGVRAQYHPHADSHVDTEENVYRFLDGTDGEFVNLCLDTGHISYCGG
Mvan_0386|M.vanbaalenii_PYR-1 FETYGVRAQYHPHADSHVDTEENVYRFLDGTDGRYVNLCLDTGHISYCGG
Mflv_2612|M.gilvum_PYR-GCK SAR-GLTAVLHPHVGTMVETRAEVDRVLAGSR---IPLCLDTGHLLIGGT
*: * ***..: *:*. :* *.* *: : *******: *
MSMEG_4665|M.smegmatis_MC2_155 DNIAIIRHAPERIGYLHLKQVDPEVRAKVEAEDLPFGEAVKLGAMTEPPR
Mvan_0386|M.vanbaalenii_PYR-1 DNIAIVRRAPERIGYLHLKQVDPEVRTKVEAEDLPFGEAVKLGAMTEPPR
Mflv_2612|M.gilvum_PYR-GCK DPLALTHEVPERIRHAHLKDVDAGLAAKVQSGELTYTQAVAAGMYVPLGA
* :*: :..**** : ***:**. : :**:: :*.: :** * .
MSMEG_4665|M.smegmatis_MC2_155 GIPDMPPLLAEIERLGVDVFAIVEQD-MYPCDPGSPLPIAQRTRSYLRSC
Mvan_0386|M.vanbaalenii_PYR-1 GIPDMPPLLAEIEKLGIEVFAIVEQD-MYPCEPDAPLPIAKRTRSYLGSC
Mflv_2612|M.gilvum_PYR-GCK GDVDIAGIITALEDNGFDGWYVMEQDKILHANPGATPDGRGPLGDVLASV
* *:. ::: :* *.: : ::*** : .:*.:. . * *
MSMEG_4665|M.smegmatis_MC2_155 GVPAVRFT----
Mvan_0386|M.vanbaalenii_PYR-1 GIPSVTFK----
Mflv_2612|M.gilvum_PYR-GCK AFLESAATGVSS
.. .