For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGLDELFAQIPVADIANKVGADEGEVTAAIKTLVPALVGGLQQNVAADDIDSAPLESAVTEQAASGLLDG GVNVDDVDEKQGDNLVAHLFGGNDSNAVASALAGTGAGNGDLIKKLLPILAPIVLAYIGKQFSQQNAPAQ QQASGGGLADILGSILGGMASGGQQANQSANNNPLGSILGSVLGGGQGNAIGEILGGLLGGKK*
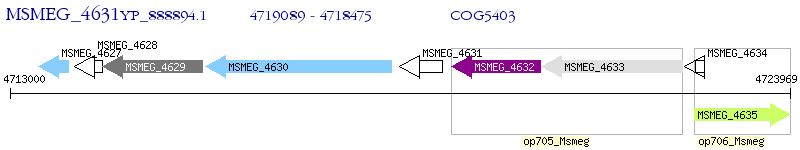
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4631 | - | - | 100% (204) | hypothetical protein MSMEG_4631 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2629 | - | 1e-47 | 51.96% (204) | hypothetical protein Mflv_2629 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1602 | - | 4e-53 | 56.94% (209) | hypothetical protein MAB_1602 |
| M. marinum M | MMAR_3774 | - | 3e-50 | 53.05% (213) | hypothetical protein MMAR_3774 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3721 | - | 4e-48 | 52.36% (212) | hypothetical protein MUL_3721 |
| M. vanbaalenii PYR-1 | Mvan_3952 | - | 2e-59 | 61.76% (204) | hypothetical protein Mvan_3952 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3774|M.marinum_M ---MAGLDDLYARIPTSEIASRLGVDQGEVDSAVHTLVPVLVSGLHQT--
MUL_3721|M.ulcerans_Agy99 MASMAGLDDLYARIPTSEIASRLGVDQGEVDSAVHVPVPVLVSGLHQT--
Mflv_2629|M.gilvum_PYR-GCK ---MTDLDDLYSEIPTQQIAQRLGADEREVGDAVKTLLPVLVGGLQHN--
Mvan_3952|M.vanbaalenii_PYR-1 ---MADLDELYGEIPTRQIAARLGADEAEVSSAVKTLLPVLVGGLQQN--
MSMEG_4631|M.smegmatis_MC2_155 ---MAGLDELFAQIPVADIANKVGADEGEVTAAIKTLVPALVGGLQQNVA
MAB_1602|M.abscessus_ATCC_1997 ---MATLDELLNEIPTDQIAAKLGVDNETADKAIKSVVPLLVGGLQAN--
*: **:* .**. :** ::*.*: . *:: :* **.**: .
MMAR_3774|M.marinum_M SQDPDHASRIESAANRQAARGLLDAGGGLDHNLDAADRGDGHQTIATLFG
MUL_3721|M.ulcerans_Agy99 SQDPDHASRIESAANRQAARGLLDAGGGLDHNLDAADRGDGHQTIATLFG
Mflv_2629|M.gilvum_PYR-GCK AADPGTASAIASAANNHAG--LLDGG----VDVEQVDEADGSKAVAKIFG
Mvan_3952|M.vanbaalenii_PYR-1 ALDPGTASTIASAANSHAG--LLDGG----VDVDQVDEVDGSKAVAKIFG
MSMEG_4631|M.smegmatis_MC2_155 ADDIDSAPLESAVTEQAAS-GLLDGG----VNVDDVDEKQGDNLVAHLFG
MAB_1602|M.abscessus_ATCC_1997 ADDPDKAADLQANVNSAPS-TLLDGG----VNVDDVDQQAGNAVVASLFG
: * . *. : .: .. ***.* ::: .*. * :* :**
MMAR_3774|M.marinum_M GNDAEQVASALAKGGAGNRDLLRQLLPIVLPIVLAYIGNQLGSG--EGSQ
MUL_3721|M.ulcerans_Agy99 GNDAEQVASALAKGGAGNRDLLKQLLPIVLPIVLAYIGNQLGSG--EGSQ
Mflv_2629|M.gilvum_PYR-GCK GNDTGQVAAALSGGGAGNSDLINKLLPILAPIVLAYLGRKLS-----GGQ
Mvan_3952|M.vanbaalenii_PYR-1 GNDTGQVASALSGGGAGNSDLINRLLPILAPIVLAYIGRKLS-----GGQ
MSMEG_4631|M.smegmatis_MC2_155 GNDSNAVASALAGTGAGNGDLIKKLLPILAPIVLAYIGKQFS---QQNAP
MAB_1602|M.abscessus_ATCC_1997 DNNADDVAAKLAGAGAGNSDLIKQLLPILTPIVLAFVAKKLTGGGAAAAP
.*:: **: *: **** **:.:****: *****::..:: .
MMAR_3774|M.marinum_M QNSESSSRGGLGEVLGSIL-----GGGS-----GDKTLGSILGSVLGG-K
MUL_3721|M.ulcerans_Agy99 QNSESASRGGLGEVLGCIL-----GGGS-----VDKSLGSILGSVLGG-K
Mflv_2629|M.gilvum_PYR-GCK STP---------------------GGG-------DNPLGSILGSVLGGDK
Mvan_3952|M.vanbaalenii_PYR-1 AQPQGQSGGGLGDILGGIL-----GGGGQRGAGADNPLGSILGSVLGGDK
MSMEG_4631|M.smegmatis_MC2_155 AQQ-QASGGGLADILGSILGGMASGGQQANQSANNNPLGSILGSVLGG-G
MAB_1602|M.abscessus_ATCC_1997 AEAPAASGGGIGDILGNILGSAVGGGAAAGG---NNPLGSILGSVLGSKG
** ::.**********.
MMAR_3774|M.marinum_M GG-ALGDILGGLLGGKK
MUL_3721|M.ulcerans_Agy99 GG-ALGDILGGLLGGKT
Mflv_2629|M.gilvum_PYR-GCK GG-PLGNILGGLLGGRK
Mvan_3952|M.vanbaalenii_PYR-1 GG-PLGDILGGLLGGKK
MSMEG_4631|M.smegmatis_MC2_155 QGNAIGEILGGLLGGKK
MAB_1602|M.abscessus_ATCC_1997 GSDALGSILGGLLGGKK
. .:*.********:.