For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSTAEPHRTDTTPALSPVAANLEAVAEKLGVKSVLVMRSEPDSMVVAATAGEATKHYTVGAAGRKAGSDA GRVPLYCERVVASDETLFVRDSRQDNTFAGNEDETEFGLSNYLGLAVHDGSGNVVGTVCVLDDTARDYSD DDRRELEELRAQVEQIVARDDTALG
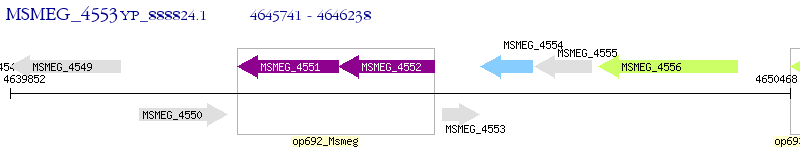
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4553 | - | - | 100% (165) | GAF domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1493 | - | 1e-07 | 29.91% (107) | putative GAF sensor protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5263 | - | 1e-05 | 31.51% (73) | putative GAF sensor protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1493|M.gilvum_PYR-GCK --MRGDLPIGLRRLAQPLGRRAGGSRRSNGPRLCRPGGRHPADGRHQAAR
Mvan_5263|M.vanbaalenii_PYR-1 MVSVSGFDDWLNRLVMELADKAG---LSPDRYVAEAVAMRLMDDVTTNDG
MSMEG_4553|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1493|M.gilvum_PYR-GCK RRDGGLLRASLRGRHRPSPGN-QRPQSRRVRPARLQALQSTGLLDAPPDV
Mvan_5263|M.vanbaalenii_PYR-1 VHD--ELLAHLSNAQIPVPAQWQWPQSIITDPERLRVLYATGLLDSPAEK
MSMEG_4553|M.smegmatis_MC2_155 ------MSTAEPHRTDTTPALSPVAANLEAVAEKLGVKSVLVMRSEP---
: : . *. . . . . :* . : . *
Mflv_1493|M.gilvum_PYR-GCK AYNRITDMAATALAAPGAAITLVDADRQFFVAMHGSATDEPEARETPLAR
Mvan_5263|M.vanbaalenii_PYR-1 VYDRIVDMAAAALAAPGAAVSLVDHDRQFFVSMHGSTGQEPESRETSIQR
MSMEG_4553|M.smegmatis_MC2_155 -----DSMVVAATAG--------EATKHYTVGAAGRKAGSDAGR----VP
.*..:* *. : ::: *. * . .*
Mflv_1493|M.gilvum_PYR-GCK SICQYAVAAGQPLVVSDARTDPVLKYNPAVTDGTVVSYLGIPLIDGSHHA
Mvan_5263|M.vanbaalenii_PYR-1 SLCQYAVASGQPLVLADARTDPVLQHNPAVSDGSVVAYLGIPLIGRDHHA
MSMEG_4553|M.smegmatis_MC2_155 LYCERVVASDETLFVRDSRQDNTFAGNEDETEFGLSNYLGLAVHDGSGNV
*: .**:.:.*.: *:* * .: * :: : ***:.: . . :.
Mflv_1493|M.gilvum_PYR-GCK IGTLCVWDTSPREWTSGHVTTLRDLAHLASARIFNE-------
Mvan_5263|M.vanbaalenii_PYR-1 VGTLCVWDNTPRDWTSGHVTTLRDLAQLAADHIFPRAQRGQAP
MSMEG_4553|M.smegmatis_MC2_155 VGTVCVLDDTARDYSDDDRRELEELRAQVEQIVARDDTALG--
:**:** * :.*:::... *.:* . :