For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GSADDRRFEVLRAIVADFVATKEPIGSKTLVERHNLGVSSATVRNDMAVLEAEGYITQPHTSSGRVPTEK GYREFVDRIDNVKPLSSSERRAILNFLESGVDLDDVLRRAVRLLAQLTRQVAIVQYPTLSTSSVRHLEVV ALTPARLLLVVITDTGRVDQRIVELGDAIDEHELSKLRDMLGQAMEGKPLAQASIAVSDLASHLNGSDRL GDAVGRAATVLVETLVEHTEERLLLGGTANLTRNTADFGGSLRSVLEALEEQVVVLRLLAAQQEAGKVTV RIGHETEAEQMAGASVVSTAYGSSGKVYGGMGVVGPTRMDYPGTIANVAAVALYIGEVLGSR*
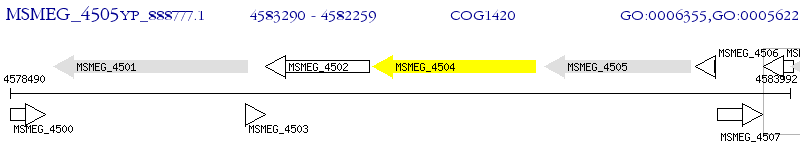
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4505 | hrcA | - | 100% (343) | heat-inducible transcription repressor |
| M. smegmatis MC2 155 | MSMEG_1377 | - | 2e-05 | 27.27% (143) | DeoR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2395c | hrcA | 1e-165 | 85.71% (343) | heat-inducible transcription repressor |
| M. gilvum PYR-GCK | Mflv_2701 | hrcA | 1e-170 | 88.92% (343) | heat-inducible transcription repressor |
| M. tuberculosis H37Rv | Rv2374c | hrcA | 1e-165 | 85.71% (343) | heat-inducible transcription repressor |
| M. leprae Br4923 | MLBr_00624 | hrcA | 1e-159 | 82.51% (343) | heat-inducible transcription repressor |
| M. abscessus ATCC 19977 | MAB_1665 | hrcA | 1e-152 | 80.17% (343) | heat-inducible transcription repressor |
| M. marinum M | MMAR_3684 | hrcA | 1e-164 | 85.13% (343) | heat shock protein transcriptional repressor HrcA |
| M. avium 104 | MAV_2022 | hrcA | 1e-165 | 85.71% (343) | heat-inducible transcription repressor |
| M. thermoresistible (build 8) | TH_1930 | hrcA | 1e-166 | 87.98% (341) | PROBABLE HEAT SHOCK PROTEIN TRANSCRIPTIONAL REPRESSOR HRCA |
| M. ulcerans Agy99 | MUL_3627 | hrcA | 1e-163 | 84.55% (343) | heat-inducible transcription repressor |
| M. vanbaalenii PYR-1 | Mvan_3837 | hrcA | 1e-170 | 88.92% (343) | heat-inducible transcription repressor |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4505|M.smegmatis_MC2_155 MG-SADDRRFEVLRAIVADFVATKEPIGSKTLVERHNLGVSSATVRNDMA
TH_1930|M.thermoresistible__bu MGSSAEERRFEVLRAIVADFVRTKEPIGSKTLVERHNLGVSSATVRNDMA
Mflv_2701|M.gilvum_PYR-GCK MG-SAEDRRFEVLRAIVADFVATKEPIGSKTLVDRHNLGVSSATVRNDMA
Mvan_3837|M.vanbaalenii_PYR-1 MG-SADDRRFEVLRAIVADFVATKEPIGSKAIVERHNLGVSSATVRNDMA
MMAR_3684|M.marinum_M MG-TADERRFEVLRAIVADFVATHEPIGSKSLVERHNLGVSSATIRNDMA
MUL_3627|M.ulcerans_Agy99 MG-TADERRFEVLRAIVADFVATHEPIGSKSLVERHNLGVSSATIRNDMA
Mb2395c|M.bovis_AF2122/97 MG-SADERRFEVLRAIVADFVATQEPIGSKSLVERHNLGVSSATVRNDMA
Rv2374c|M.tuberculosis_H37Rv MG-SADERRFEVLRAIVADFVATQEPIGSKSLVERHNLGVSSATVRNDMA
MLBr_00624|M.leprae_Br4923 MG-NADERRFEVLRAIVADFVATKEPIGSRALVERHNLGVSSATIRNDMA
MAV_2022|M.avium_104 MG-SADERRFEVLRAIVADFIATKEPIGSKTLVERHNLGVSSATVRNDMA
MAB_1665|M.abscessus_ATCC_1997 MA-SADDRRFEVLRAIVADYVTTKEPIGSKALVDRHGLGVSSATVRNDMA
*. .*::************:: *:*****:::*:**.*******:*****
MSMEG_4505|M.smegmatis_MC2_155 VLEAEGYITQPHTSSGRVPTEKGYREFVDRIDNVKPLSSSERRAILNFLE
TH_1930|M.thermoresistible__bu VLEAEGYITQPHTSSGRIPTEKGYREFVDRLDEIKPLSASERSAIQKFLE
Mflv_2701|M.gilvum_PYR-GCK VLEAEGYITQPHTSSGRVPTEKGYREFVDRLDDVKPMSSAERRAILSFLE
Mvan_3837|M.vanbaalenii_PYR-1 VLEAEGYITQPHTSSGRVPTEKGYREFVDRLQDVKPMSGAERRAILQFLE
MMAR_3684|M.marinum_M VLEAEGYIAQPHTSSGRVPTEKGYREFVDRLDDVKPLSMVERRAIQGFLE
MUL_3627|M.ulcerans_Agy99 VLEAEGYIAQPHTSSGRVPTEKGYREFVDRLDDVKPLSMVERRAIQGFLE
Mb2395c|M.bovis_AF2122/97 VLEAEGYITQPHTSSGRVPTEKGYREFVDRLEDVKPLSSAERRAIQSFLE
Rv2374c|M.tuberculosis_H37Rv VLEAEGYITQPHTSSGRVPTEKGYREFVDRLEDVKPLSSAERRAIQSFLE
MLBr_00624|M.leprae_Br4923 VLEAEGYITQPHTSSGRVPTEKGYREFVDRLEDVKPLSAAERRAIQSFLE
MAV_2022|M.avium_104 VLEAEGYITQPHTSSGRVPTEKGYREFVDRLDDVKPLSAAERRAIQNFLE
MAB_1665|M.abscessus_ATCC_1997 VLEAEGYIAQPHTSSGRVPTEKGYREFVNRLEDVKPLSGAERKAILNFLE
********:********:**********:*::::**:* ** ** ***
MSMEG_4505|M.smegmatis_MC2_155 SGVDLDDVLRRAVRLLAQLTRQVAIVQYPTLSTSSVRHLEVVALTPARLL
TH_1930|M.thermoresistible__bu SGVDLDDVLRRAVKLLAQLTRQVAIVQYPTLSTSSVRHLEVVALTPARLL
Mflv_2701|M.gilvum_PYR-GCK SGVDLDDVLRRAVRLLAQLTRQVAVVQYPMLSTSTVRRLEVVALTPARLL
Mvan_3837|M.vanbaalenii_PYR-1 SGVDLDDVLRRAVRLLAQLTRQVAVVQYPTLTTSTVRRLEVVALTPARLL
MMAR_3684|M.marinum_M SGVDLDDVLRRAVRLLAQLTRQVAVVQYPTLSTSKVRHLEVIALTPARLL
MUL_3627|M.ulcerans_Agy99 SGVDLDDVLRRAVRLLAQLTRQVAVVQYPTLSTSKVRHLEVIALTPARLL
Mb2395c|M.bovis_AF2122/97 SGVDLDDVLRRAVRLLAQLTRQVAVVQYPTLSTSTVRHLEVIALTPARLL
Rv2374c|M.tuberculosis_H37Rv SGVDLDDVLRRAVRLLAQLTRQVAVVQYPTLSTSTVRHLEVIALTPARLL
MLBr_00624|M.leprae_Br4923 SGVDLDDVLRRAVRLLAQLTCQVAVVQYPTLSTSTVRHLEVIALSPARLL
MAV_2022|M.avium_104 SGVDLDDVLRRAVRLLAQLTRQVAIVQYPTLSSSTVRHLEVIALTPARLL
MAB_1665|M.abscessus_ATCC_1997 GGVDLDDVLRRAVRLLAQMTRQVAVVQYPTLSSSSVRHLEVVSLSPARLL
.************:****:* ***:**** *::*.**:***::*:*****
MSMEG_4505|M.smegmatis_MC2_155 LVVITDTGRVDQRIVELGDAIDEHELSKLRDMLGQAMEGKPLAQASIAVS
TH_1930|M.thermoresistible__bu LVVITDTGRVDQRIVELGDPIDEHDLSRLREVLGGALEGKPLAAASAAVS
Mflv_2701|M.gilvum_PYR-GCK LIVITDSGRIDQRIVELGDAIDEDELTQLRDLLGQALEGKPLTTASIAVS
Mvan_3837|M.vanbaalenii_PYR-1 LVVITDTGRVDQRIVELGDGIDDGQLAQLRDMLGSALEGKPLAAASVAVS
MMAR_3684|M.marinum_M MVVITDSGRVDQRIVELGDVIDDHQLSQLRELLGQALEGKKLAAASVAVA
MUL_3627|M.ulcerans_Agy99 VVVITDSGRVDQRIVELGDVIDDHQLSQLRELLGQALEGKKLAAASVAVA
Mb2395c|M.bovis_AF2122/97 MVVITDSGRVDQRIVELGDVIDDHQLAQLREILGQALEGKKLSAASVAVA
Rv2374c|M.tuberculosis_H37Rv MVVITDSGRVDQRIVELGDVIDDHQLAQLREILGQALEGKKLSAASVAVA
MLBr_00624|M.leprae_Br4923 MVVITESGRVDQRIAELGDVIDDYQLSQLREMLSQAMGGKKLSAASAAVA
MAV_2022|M.avium_104 MVVITDSGRVDQRIVELGDVIDDHELSRLREMLGQALVGKKLSAASVAVA
MAB_1665|M.abscessus_ATCC_1997 LVVITDTGRVDQRIVELGDVINDEQLGRLRVLLGAALDGKKLSAASVAVA
::***::**:****.**** *:: :* :** :*. *: ** *: ** **:
MSMEG_4505|M.smegmatis_MC2_155 DLASHLNGSDRLGDAVGRAATVLVETLVEHTEERLLLGGTANLTRNTADF
TH_1930|M.thermoresistible__bu DLVAQLGGQGRFSDAVGRASTVLVETLVEHHEERLLLGGTANLTRNTADF
Mflv_2701|M.gilvum_PYR-GCK DLASHLGGQGRLANAVGRSATVLVESLVEHREERLLLGGTANLTRNTADF
Mvan_3837|M.vanbaalenii_PYR-1 DLAVHLNGQGGLADAVGRSATVLVETLVEHTEERLLLGGTANLTRNTADF
MMAR_3684|M.marinum_M DLAGQLSGAGGLGDAVGRSATVLLESLVEHTEERLLMGGTANLTRNAADF
MUL_3627|M.ulcerans_Agy99 DFAGQLSGAGGLGDAVGRSATVLLESLVEHTEERLLMGGTANLTRNAADF
Mb2395c|M.bovis_AF2122/97 DLASQLGGAGGLGDAVGRAATVLLESLVEHTEERLLLGGTANLTRNAADF
Rv2374c|M.tuberculosis_H37Rv DLASQLGGAGGLGDAVGRAATVLLESLVEHTEERLLLGGTANLTRNAADF
MLBr_00624|M.leprae_Br4923 DLVGRFRGTGGLANAVGRSANVLLESLVEHTEERLLLGGTANLTRNSADF
MAV_2022|M.avium_104 DLAEQLRSPDGLGDAVGRSATVLLESLVEHSEERLLMGGTANLTRNAADF
MAB_1665|M.abscessus_ATCC_1997 ELAEQSD--DDLRDPITRAATVLVETLVEHHEERLLLGGTANLTRNAADF
::. : . : :.: *::.**:*:**** *****:*********:***
MSMEG_4505|M.smegmatis_MC2_155 GGSLRSVLEALEEQVVVLRLLAAQQEAGKVTVRIGHETEAEQMAGASVVS
TH_1930|M.thermoresistible__bu GGSLRSVLEALEEQVVVLKLLDKQQQAGKVTVRIGHETEAEEMASASVVT
Mflv_2701|M.gilvum_PYR-GCK GGSLRSLLEALEEQVVVLRLLAKQQEAGKVTVRIGHETEAEEMAGASVVT
Mvan_3837|M.vanbaalenii_PYR-1 GGSLRSVLEALEEQVVVLRLLAKQQEAGKVTVRIGHETEAEEMAGTSVIS
MMAR_3684|M.marinum_M GGSLRSILEALEEQVVVLRLLAAQQEAGKVTVRIGHETEAEQIVGTSMVS
MUL_3627|M.ulcerans_Agy99 GGSLRSILEALEEQIVVLRLLAAQQEAGKVTVRIGHETEAEQIVGTSMVS
Mb2395c|M.bovis_AF2122/97 GGSLRSILEALEEQVVVLRLLAAQQEAGKVTVRIGHETASEQMVGTSMVS
Rv2374c|M.tuberculosis_H37Rv GGSLRSILEALEEQVVVLRLLAAQQEAGKVTVRIGHETASEQMVGTSMVS
MLBr_00624|M.leprae_Br4923 GGSLRSILEALEEQVVVLRLLAAQQEAGKVTVRIGHETAAEQIMGTSMVS
MAV_2022|M.avium_104 GGSLRSILEALEEQVVVLRLLAAQQEAGKVTVRIGYETAAEQMVGTSMVT
MAB_1665|M.abscessus_ATCC_1997 GGQLRTVLEALEEQVVVLRLLAAQQEAGRVTVHIGHETAAEQMIGTSVVS
**.**::*******:***:** **:**:***:**:** :*:: .:*:::
MSMEG_4505|M.smegmatis_MC2_155 TAYGSSGKVYGGMGVVGPTRMDYPGTIANVAAVALYIGEVLGSR
TH_1930|M.thermoresistible__bu TAYGSAGKVYGGMGVVGPTRMDYPGTMANVAAVALYIGEVLGNR
Mflv_2701|M.gilvum_PYR-GCK TAYGSAGKVFGGMGVLGPTRMDYPGTIANVAAVALYIGEVLGNR
Mvan_3837|M.vanbaalenii_PYR-1 TAYGSAGKVFGGMGVLGPTRMDYPGTIANVAAVALYIGEVLGNR
MMAR_3684|M.marinum_M TAYGSNDTVYGGMGVLGPTRMDYPGTIASVAAVALYIGEVLGAR
MUL_3627|M.ulcerans_Agy99 TAYGSNDTVYGGMGVLGPTRMDYPGTIASVAAVALYIGEVLGAR
Mb2395c|M.bovis_AF2122/97 TAYGTAHTVYGGMGVVGPTRMDYPGTIASVAAVALYIGDVLGAR
Rv2374c|M.tuberculosis_H37Rv TAYGTAHTVYGGMGVVGPTRMDYPGTIASVAAVALYIGDVLGAR
MLBr_00624|M.leprae_Br4923 TAYGTSGTVYGGMGVLGPTRMDYPGTIASVAAVALYIGEVLGAR
MAV_2022|M.avium_104 TAYGTSDTVYGGMGVLGPTRMDYPGTIASVAAVAMYIGEVLGAR
MAB_1665|M.abscessus_ATCC_1997 TPYGAGGAVFGGMGVLGPTRMDYPGTIANVAAVAMYIGEVLANR
*.**: *:*****:**********:*.*****:***:**. *