For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTTGATRTWFPDERHAEVLRLLDAEHRVESAQLASLFGVSAECVRKDLAQLEARGLLRRVHGGAVPAIS SRTEPDVADRIENAEAKDAIARHALRYVSEGATLLLDAGSTTQRLAEMLSTAAELVVYTNAVPVATTLLR RGITAVLLGGRIREATMAAVGALTTEALAAINVDVAFLGTNALSLDRGLTTPDPEEAAVKRHMLAAAGQR VFLVDSSKFGRHSQARHAELSDVDVLITDDGADPGLRGQLRAAGITVEVAHR
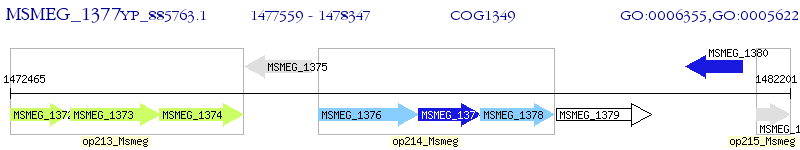
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1377 | - | - | 100% (262) | DeoR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_0087 | - | 7e-49 | 44.88% (254) | glucitol operon repressor |
| M. smegmatis MC2 155 | MSMEG_0509 | - | 2e-27 | 34.14% (249) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2395c | hrcA | 6e-06 | 26.57% (143) | heat-inducible transcription repressor |
| M. gilvum PYR-GCK | Mflv_0750 | - | 2e-51 | 47.41% (251) | DeoR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv2374c | hrcA | 6e-06 | 26.57% (143) | heat-inducible transcription repressor |
| M. leprae Br4923 | MLBr_00624 | hrcA | 2e-06 | 26.57% (143) | heat-inducible transcription repressor |
| M. abscessus ATCC 19977 | MAB_1665 | hrcA | 5e-05 | 26.02% (123) | heat-inducible transcription repressor |
| M. marinum M | MMAR_3684 | hrcA | 4e-05 | 26.02% (123) | heat shock protein transcriptional repressor HrcA |
| M. avium 104 | MAV_2022 | hrcA | 8e-06 | 27.64% (123) | heat-inducible transcription repressor |
| M. thermoresistible (build 8) | TH_3302 | - | 5e-30 | 36.96% (230) | PUTATIVE putative glucitol operon repressor |
| M. ulcerans Agy99 | MUL_3627 | hrcA | 2e-05 | 26.02% (123) | heat-inducible transcription repressor |
| M. vanbaalenii PYR-1 | Mvan_0096 | - | 4e-48 | 46.78% (233) | DeoR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2395c|M.bovis_AF2122/97 --------MGSADERRFEVLRAIVADFVATQEPIGSKSLVERHNLGVSSA
Rv2374c|M.tuberculosis_H37Rv --------MGSADERRFEVLRAIVADFVATQEPIGSKSLVERHNLGVSSA
MMAR_3684|M.marinum_M --------MGTADERRFEVLRAIVADFVATHEPIGSKSLVERHNLGVSSA
MUL_3627|M.ulcerans_Agy99 --------MGTADERRFEVLRAIVADFVATHEPIGSKSLVERHNLGVSSA
MAV_2022|M.avium_104 --------MGSADERRFEVLRAIVADFIATKEPIGSKTLVERHNLGVSSA
MLBr_00624|M.leprae_Br4923 --------MGNADERRFEVLRAIVADFVATKEPIGSRALVERHNLGVSSA
MAB_1665|M.abscessus_ATCC_1997 --------MASADDRRFEVLRAIVADYVTTKEPIGSKALVDRHGLGVSSA
Mflv_0750|M.gilvum_PYR-GCK ---------MYPEERQQAIASLVMTKGRASVTELAH-------AYDVTTE
Mvan_0096|M.vanbaalenii_PYR-1 ---------MYPEERQQAIASLVMSKGRASVTELAQ-------AYDVTTE
MSMEG_1377|M.smegmatis_MC2_155 MTTTGATRTWFPDERHAEVLRLLDAEHRVESAQLAS-------LFGVSAE
TH_3302|M.thermoresistible__bu ---------LLASDRRREIFNRLVREGYVEAKELAE-------ELGVDAS
..:*: : : . . :. .* :
Mb2395c|M.bovis_AF2122/97 TVRNDMAVLEAEGYITQPHTSSGRVPTEKGYREFVDRLEDVKPLSSAERR
Rv2374c|M.tuberculosis_H37Rv TVRNDMAVLEAEGYITQPHTSSGRVPTEKGYREFVDRLEDVKPLSSAERR
MMAR_3684|M.marinum_M TIRNDMAVLEAEGYIAQPHTSSGRVPTEKGYREFVDRLDDVKPLSMVERR
MUL_3627|M.ulcerans_Agy99 TIRNDMAVLEAEGYIAQPHTSSGRVPTEKGYREFVDRLDDVKPLSMVERR
MAV_2022|M.avium_104 TVRNDMAVLEAEGYITQPHTSSGRVPTEKGYREFVDRLDDVKPLSAAERR
MLBr_00624|M.leprae_Br4923 TIRNDMAVLEAEGYITQPHTSSGRVPTEKGYREFVDRLEDVKPLSAAERR
MAB_1665|M.abscessus_ATCC_1997 TVRNDMAVLEAEGYIAQPHTSSGRVPTEKGYREFVNRLEDVKPLSGAERK
Mflv_0750|M.gilvum_PYR-GCK TVRRDLAVLDKAGIVRRVHG--GAVP--------------ARSLHLVEPG
Mvan_0096|M.vanbaalenii_PYR-1 TVRRDLAVLDKAGVVRRVHG--GAVP--------------VRALHLVEPG
MSMEG_1377|M.smegmatis_MC2_155 CVRKDLAQLEARGLLRRVHG--GAVP--------------AISSR-TEPD
TH_3302|M.thermoresistible__bu TIRRDLDTLARSGQVKRTHG--GART--------------VPGVS-ADLP
:*.*: * * : : * * . . .:
Mb2395c|M.bovis_AF2122/97 AIQSFLESGVDLDDVLRRAVRLLAQLTRQVAVVQYPTLSTSTVRHLEVIA
Rv2374c|M.tuberculosis_H37Rv AIQSFLESGVDLDDVLRRAVRLLAQLTRQVAVVQYPTLSTSTVRHLEVIA
MMAR_3684|M.marinum_M AIQGFLESGVDLDDVLRRAVRLLAQLTRQVAVVQYPTLSTSKVRHLEVIA
MUL_3627|M.ulcerans_Agy99 AIQGFLESGVDLDDVLRRAVRLLAQLTRQVAVVQYPTLSTSKVRHLEVIA
MAV_2022|M.avium_104 AIQNFLESGVDLDDVLRRAVRLLAQLTRQVAIVQYPTLSSSTVRHLEVIA
MLBr_00624|M.leprae_Br4923 AIQSFLESGVDLDDVLRRAVRLLAQLTCQVAVVQYPTLSTSTVRHLEVIA
MAB_1665|M.abscessus_ATCC_1997 AILNFLEGGVDLDDVLRRAVRLLAQMTRQVAVVQYPTLSSSSVRHLEVVS
Mflv_0750|M.gilvum_PYR-GCK VGERDVTRTEHKDAIAAAAVEFLPLSGATVLLDAGTTT-------MRIAG
Mvan_0096|M.vanbaalenii_PYR-1 VGERDVIRAEHKDAIAAAAAEFFPLSGASVLLDAGTTT-------MRIAA
MSMEG_1377|M.smegmatis_MC2_155 VADR-IENAEAKDAIARHALRYVS-EGATLLLDAGSTT-------QRLAE
TH_3302|M.thermoresistible__bu YSMKETARLDQKAAIAGVAVQRVR-DGDTVVLDSGSTT-------YEIAV
: * . . : : .* .:
Mb2395c|M.bovis_AF2122/97 LTPARLLMVVITDSGRVDQRIVELGDVIDDHQLAQLREILGQALEGKKLS
Rv2374c|M.tuberculosis_H37Rv LTPARLLMVVITDSGRVDQRIVELGDVIDDHQLAQLREILGQALEGKKLS
MMAR_3684|M.marinum_M LTPARLLMVVITDSGRVDQRIVELGDVIDDHQLSQLRELLGQALEGKKLA
MUL_3627|M.ulcerans_Agy99 LTPARLLVVVITDSGRVDQRIVELGDVIDDHQLSQLRELLGQALEGKKLA
MAV_2022|M.avium_104 LTPARLLMVVITDSGRVDQRIVELGDVIDDHELSRLREMLGQALVGKKLS
MLBr_00624|M.leprae_Br4923 LSPARLLMVVITESGRVDQRIAELGDVIDDYQLSQLREMLSQAMGGKKLS
MAB_1665|M.abscessus_ATCC_1997 LSPARLLLVVITDTGRVDQRIVELGDVINDEQLGRLRVLLGAALDGKKLS
Mflv_0750|M.gilvum_PYR-GCK ELPADRELVVVTNSVPIAARLAAMPSISLQLLGGRVRGVTQAAVGEQTLR
Mvan_0096|M.vanbaalenii_PYR-1 QLPADRELVIVTNSVPIAARLATVPSVSLQLLGGRVRGVTQAAVGEQALR
MSMEG_1377|M.smegmatis_MC2_155 MLSTAAELVVYTNAVPVATTLLRR-GITAVLLGGRIREATMAAVGALTTE
TH_3302|M.thermoresistible__bu RLRNHEDLTVITNDLRIATFIADLRRFRLLVAGGELLGSVYTLTGRRTVE
:.: *: : : . ..:
Mb2395c|M.bovis_AF2122/97 AASVAVADLASQLGGAGGLGDAVGRAATVLLESLVEHTEERLLLGGTANL
Rv2374c|M.tuberculosis_H37Rv AASVAVADLASQLGGAGGLGDAVGRAATVLLESLVEHTEERLLLGGTANL
MMAR_3684|M.marinum_M AASVAVADLAGQLSGAGGLGDAVGRSATVLLESLVEHTEERLLMGGTANL
MUL_3627|M.ulcerans_Agy99 AASVAVADFAGQLSGAGGLGDAVGRSATVLLESLVEHTEERLLMGGTANL
MAV_2022|M.avium_104 AASVAVADLAEQLRSPDGLGDAVGRSATVLLESLVEHSEERLLMGGTANL
MLBr_00624|M.leprae_Br4923 AASAAVADLVGRFRGTGGLANAVGRSANVLLESLVEHTEERLLLGGTANL
MAB_1665|M.abscessus_ATCC_1997 AASVAVAELAEQSD--DDLRDPITRAATVLVETLVEHHEERLLLGGTANL
Mflv_0750|M.gilvum_PYR-GCK VLDTLRVDIA--FIGTNAISARHGLSTPDSEEAAVKR--AMVKAANYVVV
Mvan_0096|M.vanbaalenii_PYR-1 VLDTLRVDIA--FIGTNAISVRHGLSTPDSEEAAVKR--AMVRAANYVVV
MSMEG_1377|M.smegmatis_MC2_155 ALAAINVDVA--FLGTNALSLDRGLTTPDPEEAAVKR--HMLAAAGQRVF
TH_3302|M.thermoresistible__bu FLLDYAADWT--FLGADAVDAAAGITNTNTLEIPVKR--AMLEVAATTVV
.: . . : : * *:: : . .
Mb2395c|M.bovis_AF2122/97 TRNAADFGGSLRSILEALEEQVVVLRLLAAQQEAGKVTVRIGHETASEQM
Rv2374c|M.tuberculosis_H37Rv TRNAADFGGSLRSILEALEEQVVVLRLLAAQQEAGKVTVRIGHETASEQM
MMAR_3684|M.marinum_M TRNAADFGGSLRSILEALEEQVVVLRLLAAQQEAGKVTVRIGHETEAEQI
MUL_3627|M.ulcerans_Agy99 TRNAADFGGSLRSILEALEEQIVVLRLLAAQQEAGKVTVRIGHETEAEQI
MAV_2022|M.avium_104 TRNAADFGGSLRSILEALEEQVVVLRLLAAQQEAGKVTVRIGYETAAEQM
MLBr_00624|M.leprae_Br4923 TRNSADFGGSLRSILEALEEQVVVLRLLAAQQEAGKVTVRIGHETAAEQI
MAB_1665|M.abscessus_ATCC_1997 TRNAADFGGQLRTVLEALEEQVVVLRLLAAQQEAGRVTVHIGHETAAEQM
Mflv_0750|M.gilvum_PYR-GCK AADSSKVGRED-------------FVSFAPISSVDSLITDPEISPTDSAE
Mvan_0096|M.vanbaalenii_PYR-1 AADSSKVGRED-------------FVSFAPISSVDTLITDPEISAADSAE
MSMEG_1377|M.smegmatis_MC2_155 LVDSSKFGRHS-------------QARHAELSDVDVLITDDGADPGLRGQ
TH_3302|M.thermoresistible__bu VADSSKFGHRA-------------LAKVADLDEVDEIITDSDLPSDSVDV
:::..* * .... : . .
Mb2395c|M.bovis_AF2122/97 VGTSMVSTAYGTAHTVYGGMGVVGPTRMDYPGTIASVAAVALYIGDVLGA
Rv2374c|M.tuberculosis_H37Rv VGTSMVSTAYGTAHTVYGGMGVVGPTRMDYPGTIASVAAVALYIGDVLGA
MMAR_3684|M.marinum_M VGTSMVSTAYGSNDTVYGGMGVLGPTRMDYPGTIASVAAVALYIGEVLGA
MUL_3627|M.ulcerans_Agy99 VGTSMVSTAYGSNDTVYGGMGVLGPTRMDYPGTIASVAAVALYIGEVLGA
MAV_2022|M.avium_104 VGTSMVTTAYGTSDTVYGGMGVLGPTRMDYPGTIASVAAVAMYIGEVLGA
MLBr_00624|M.leprae_Br4923 MGTSMVSTAYGTSGTVYGGMGVLGPTRMDYPGTIASVAAVALYIGEVLGA
MAB_1665|M.abscessus_ATCC_1997 IGTSVVSTPYGAGGAVFGGMGVLGPTRMDYPGTIANVAAVAMYIGEVLAN
Mflv_0750|M.gilvum_PYR-GCK LTAHGVEVICAGGRS-----------------------------------
Mvan_0096|M.vanbaalenii_PYR-1 FGEHGVEVICAGGRP-----------------------------------
MSMEG_1377|M.smegmatis_MC2_155 LRAAGITVEVAHR-------------------------------------
TH_3302|M.thermoresistible__bu FGKNLITVADPAPTPG----------------------------------
. : .
Mb2395c|M.bovis_AF2122/97 R
Rv2374c|M.tuberculosis_H37Rv R
MMAR_3684|M.marinum_M R
MUL_3627|M.ulcerans_Agy99 R
MAV_2022|M.avium_104 R
MLBr_00624|M.leprae_Br4923 R
MAB_1665|M.abscessus_ATCC_1997 R
Mflv_0750|M.gilvum_PYR-GCK -
Mvan_0096|M.vanbaalenii_PYR-1 -
MSMEG_1377|M.smegmatis_MC2_155 -
TH_3302|M.thermoresistible__bu -