For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAKTRNTAETKAAKAAAKAARKEASKQRRSQLWQAFQIQRKEDKRLVPYMAGAFVLIVAIAVVAGLLIGG FTKFTLIPLGVVLGALVAFIIFGRRAQKSVYRKAEGQTGAAAWALDNLRGKWRVTPGVAATGHFDAVHRV IGRPGVIFVGEGAPNRVKPLLAQEKKRTARLVGETPIYDIIVGNGEGEVPLAKLERHLNKLPANITVKQM DAIESRLAALGTKVGPAAVPKGPLPQAAKMRGVQRTARRR
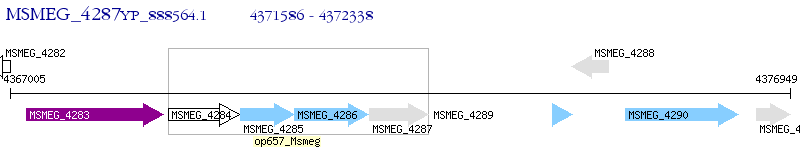
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4287 | - | - | 100% (250) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2242 | - | 1e-112 | 78.80% (250) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2931 | - | 1e-119 | 86.67% (240) | hypothetical protein Mflv_2931 |
| M. tuberculosis H37Rv | Rv2219 | - | 1e-113 | 79.20% (250) | transmembrane protein |
| M. leprae Br4923 | MLBr_00857 | - | 1e-108 | 76.80% (250) | hypothetical protein MLBr_00857 |
| M. abscessus ATCC 19977 | MAB_1940c | - | 2e-97 | 72.11% (251) | hypothetical protein MAB_1940c |
| M. marinum M | MMAR_3287 | - | 1e-111 | 77.20% (250) | transmembrane protein |
| M. avium 104 | MAV_2269 | - | 1e-113 | 78.40% (250) | integral membrane protein |
| M. thermoresistible (build 8) | TH_0218 | - | 1e-113 | 79.68% (251) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_1342 | - | 1e-112 | 77.20% (250) | transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_3583 | - | 1e-124 | 87.20% (250) | hypothetical protein Mvan_3583 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2242|M.bovis_AF2122/97 MAKPRNAAESKAAKAQANAARKAAARQRRAQLWQAFTLQRKEDKRLLPYM
Rv2219|M.tuberculosis_H37Rv MAKPRNAAESKAAKAQANAARKAAARQRRAQLWQAFTLQRKEDKRLLPYM
MMAR_3287|M.marinum_M MARSRNSAENKAAKAEAKAARKAAARERRTQLWQAFNLQRKEDKRLLPYM
MUL_1342|M.ulcerans_Agy99 MARSRNSAENKAAKAEAKAARKAAARERRTQLWQAFNLQRKEDKRLLPYM
MAV_2269|M.avium_104 MAKSRSAADNKAARAQAQAARKAAARERRAQLWQAFNIQRQEDKRLLPYM
MLBr_00857|M.leprae_Br4923 MAKPRNAAAHKAARAEAKAARKAASRQRRLQLWQAFTIQRTEDKRLIPYM
TH_0218|M.thermoresistible__bu MVKSRNSAAAKAAKAEAKAARKAAARQRRSQLWQAFKIQRQEDKRLLPYM
MSMEG_4287|M.smegmatis_MC2_155 MAKTRNTAETKAAKAAAKAARKEASKQRRSQLWQAFQIQRKEDKRLVPYM
Mflv_2931|M.gilvum_PYR-GCK MAN-------RAAKKEAKAARKAASKQRRSQLWQAFQIQRKEDKRLLPYM
Mvan_3583|M.vanbaalenii_PYR-1 MAKSRNPAETKAAKAEAKAARKAASKQRRSQLWQAFQMQRKEDKRLLPYM
MAB_1940c|M.abscessus_ATCC_199 MAKSLSPAEAKAAKAEAKARRKAESKARRAQMWQAFQIQRKEDKRLLPYM
*.. :**: *:* ** :: ** *:**** :** *****:***
Mb2242|M.bovis_AF2122/97 IGAFLLIVGASVGVGVWAGGFTMLTMIPLGVLLGALVAFVIFGRRAQRTV
Rv2219|M.tuberculosis_H37Rv IGAFLLIVGASVGVGVWAGGFTMFTMIPLGVLLGALVAFVIFGRRAQRTV
MMAR_3287|M.marinum_M IGAFVLIVGISVAVGVWAGGLTMITMIPFGLLLGGLVAFIIFGRRAQRAV
MUL_1342|M.ulcerans_Agy99 IGAFVLIVGISVAVGVWAGGLTMITMIPFGLLLGGLVAFIIFGRRAQRAV
MAV_2269|M.avium_104 IGAFLLVVGVSVAVGVWAGGLTMITLIPFGVVLGALVAFIVFGRRAQKSV
MLBr_00857|M.leprae_Br4923 IAAFSLMVSASVTAGVLVGGLTMITLILLGVVLGALVAFIIFGRRTQQSV
TH_0218|M.thermoresistible__bu ISAFVLIVAASVVLGVLAGGFTMYMLIPLGVVLGALVAFIIFGRRAQRTV
MSMEG_4287|M.smegmatis_MC2_155 AGAFVLIVAIAVVAGLLIGGFTKFTLIPLGVVLGALVAFIIFGRRAQKSV
Mflv_2931|M.gilvum_PYR-GCK IGAFVLIVGLSVVGGIYAGGFTTYMLIPLGIVLGALVAFIIFGRRAQKSV
Mvan_3583|M.vanbaalenii_PYR-1 IGAFVLIVAASVAVGLYAGGFTMFMLIPLGIVLGALVAFIIFGRRAQKSV
MAB_1940c|M.abscessus_ATCC_199 IGAIVVIVAAFAVAGYFSGGISTFLFPILGVVLGVLVAFIIFGRRVQKSV
.*: ::*. . * **:: : :*::** ****::****.*::*
Mb2242|M.bovis_AF2122/97 YRKAEGQTGAAAWALDNLRGKWRVTPGVAATGNLDAVHRVIGRPGVIFVG
Rv2219|M.tuberculosis_H37Rv YRKAEGQTGAAAWALDNLRGKWRVTPGVAATGNLDAVHRVIGRPGVIFVG
MMAR_3287|M.marinum_M YSKAEGQTGAAAWALENLRGKWRVSPGVAATGHFDAVHRVLGRPGVILVG
MUL_1342|M.ulcerans_Agy99 YSKAEGQTGAAAWALENLRGKWRVSPGVAATGHFDAVHRVLGRPGVILVG
MAV_2269|M.avium_104 YRKAEGQTGAAAWALDNLRGKWRVTPGVAATGHFDAVHRVIGRPGVILVG
MLBr_00857|M.leprae_Br4923 YHKAEGQTGGAAWALDNLRGKWRVSPGVAANGHFDAVHRVIGRPGVIFVA
TH_0218|M.thermoresistible__bu YQKAEGQTGAAAWALENMRGKWRVTPGVAATGHFDAVHRVIGRPGVILVG
MSMEG_4287|M.smegmatis_MC2_155 YRKAEGQTGAAAWALDNLRGKWRVTPGVAATGHFDAVHRVIGRPGVIFVG
Mflv_2931|M.gilvum_PYR-GCK YKKAEGQTGAAAWALDNMRGKWRVTPGVAATGHFDAVHRVIGRPGVIFVG
Mvan_3583|M.vanbaalenii_PYR-1 YRKAEGQTGAAAWALDNMRGKWRVTPGVAATGHFDAVHRVIGRPGVIFVA
MAB_1940c|M.abscessus_ATCC_199 FTKAEGQKGAAGWALDNMRGRWRVTTAVAGTTQLDVVHRVIGRPGIIFVA
: *****.*.*.***:*:**:***:..**.. ::*.****:****:*:*.
Mb2242|M.bovis_AF2122/97 EGSAARVKPLLAQEKKRTARLVGDVPIYDIIVGNGDGEVPLAKLERHLTR
Rv2219|M.tuberculosis_H37Rv EGSAARVKPLLAQEKKRTARLVGDVPIYDIIVGNGDGEVPLAKLERHLTR
MMAR_3287|M.marinum_M EGSAARVKPLLAQEKKRTARLVGEVPIYDIIVGNGDGEIPLAKLERHLTR
MUL_1342|M.ulcerans_Agy99 EGSAARVKPLLAQEKKRTARLVGEVPIYDIIVGNGDGEIPLAKLERHLTR
MAV_2269|M.avium_104 EGSPTRVRPLLAQEKKRTARLIGDVPIYDIIVGNGEDEVPLAKLERHLTR
MLBr_00857|M.leprae_Br4923 EGSAARVKPLLAQEKKRTARLVGDVPIYDIIVGNGDGEVALVKLERHLAR
TH_0218|M.thermoresistible__bu EGSPTRLKPLLAQEKKRTARLVGDVPIYDIVVGNDEGQVPLAKLERHLTR
MSMEG_4287|M.smegmatis_MC2_155 EGAPNRVKPLLAQEKKRTARLVGETPIYDIIVGNGEGEVPLAKLERHLNK
Mflv_2931|M.gilvum_PYR-GCK EGAANRVKPLLAQEKKRTARLIGDTPIYDIIIGNGEGEVPLSKLERHLNK
Mvan_3583|M.vanbaalenii_PYR-1 EGSPTRVKPLLAQEKKRTARLVGDVPIYDVIIGNGEGEVPLAKLERHLTK
MAB_1940c|M.abscessus_ATCC_199 EGAPGRLGPLLAQEKKRTARLVGDTPIYDVIVGNEDGQVPLSKLERHLTK
**:. *: *************:*:.****:::** :.::.* ****** :
Mb2242|M.bovis_AF2122/97 LPANITVKQMDTVESRLAALGSRAG-AGVMPKGPLPTTAKMRS-VQRTVR
Rv2219|M.tuberculosis_H37Rv LPANITVKQMDTVESRLAALGSRAG-AGVMPKGPLPTTAKMRS-VQRTVR
MMAR_3287|M.marinum_M LPANITVKQLDSLESRLAALGSRAG-AGALPKGPLPNAGKMRG-VQRSVR
MUL_1342|M.ulcerans_Agy99 LPANITVKQLDSLESRLAALGSRAG-AGALPKGPLPNAGKMRG-VQRSVR
MAV_2269|M.avium_104 LPANITVKQMDTLESRLAALGSRAG-AAVMPKGPLPNAGKMRG-VQRTVR
MLBr_00857|M.leprae_Br4923 LPANISVKQVDILESRLAALGSRAG-ASLIPKGPLPNAGKMRG-VQRTVR
TH_0218|M.thermoresistible__bu LPSNISVKQMDALESRLAALSAKAGPAAAMPKGPLPSQAKLRG-MQRTVR
MSMEG_4287|M.smegmatis_MC2_155 LPANITVKQMDAIESRLAALGTKVG-PAAVPKGPLPQAAKMRG-VQRTAR
Mflv_2931|M.gilvum_PYR-GCK LPANITVKQLDALESRLAALGSKIG-PAAMPKGPLPAQAKMRG-VQRTVR
Mvan_3583|M.vanbaalenii_PYR-1 LPANITVKQMDSLESRLAALGTKVG-PAAMPKGPLPAQAKMRG-IQRTVR
MAB_1940c|M.abscessus_ATCC_199 LPANITAKQMDSLESRLAALGSRAA---AMPKGPLPGQAKMRGGMQRTIR
**:**:.**:* :*******.:: . :****** .*:*. :**: *
Mb2242|M.bovis_AF2122/97 RK
Rv2219|M.tuberculosis_H37Rv RK
MMAR_3287|M.marinum_M RR
MUL_1342|M.ulcerans_Agy99 RR
MAV_2269|M.avium_104 RK
MLBr_00857|M.leprae_Br4923 RK
TH_0218|M.thermoresistible__bu RR
MSMEG_4287|M.smegmatis_MC2_155 RR
Mflv_2931|M.gilvum_PYR-GCK RR
Mvan_3583|M.vanbaalenii_PYR-1 RR
MAB_1940c|M.abscessus_ATCC_199 RR
*: