For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVVPEGRKLLRLEVRNAQTPIERKPPWIKTRAKMGPEYTELKGLVRREGLHTVCEEAGCPNIFECWEDR EATFLIGGEQCTRRCDFCQIDTGKPADLDRDEPRRVAESVQAMGLRYSTVTGVARDDLPDGGAWLYAETV RYIKRLNPNTGVELLIPDFNGNPEQLEEVFESRPEVLAHNVETVPRIFKRIRPAFRYDRSLAVITAARNF GLVTKSNLILGMGETIDEVRTALRDLHDAGCDIITITQYLRPSPRHHPVERWVHPDEFVELSAYAEGLGF AGVLAGPLVRSSYRAGKLYAQAARVRFADQPPVS
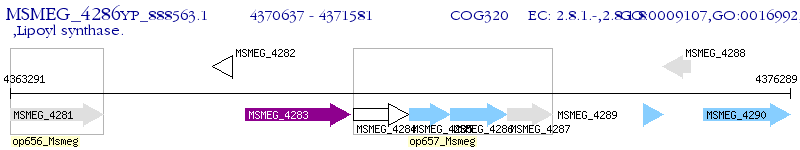
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4286 | lipA | - | 100% (314) | lipoyl synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2241 | lipA | 1e-154 | 84.64% (306) | lipoyl synthase |
| M. gilvum PYR-GCK | Mflv_2932 | - | 1e-162 | 88.64% (308) | lipoyl synthase |
| M. tuberculosis H37Rv | Rv2218 | lipA | 1e-154 | 84.64% (306) | lipoyl synthase |
| M. leprae Br4923 | MLBr_00858 | lipA | 1e-152 | 84.31% (306) | lipoyl synthase |
| M. abscessus ATCC 19977 | MAB_1942c | - | 1e-152 | 87.21% (297) | lipoyl synthase |
| M. marinum M | MMAR_3286 | lipA | 1e-158 | 88.93% (298) | lipoate biosynthesis protein a LipA |
| M. avium 104 | MAV_2270 | lipA | 1e-155 | 85.95% (306) | lipoyl synthase |
| M. thermoresistible (build 8) | TH_0219 | lipA | 1e-165 | 88.85% (314) | Probable lipoate biosynthesis protein A LipA |
| M. ulcerans Agy99 | MUL_1343 | lipA | 1e-158 | 88.93% (298) | lipoyl synthase |
| M. vanbaalenii PYR-1 | Mvan_3582 | - | 1e-159 | 86.77% (310) | lipoyl synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2932|M.gilvum_PYR-GCK MSIAP----------DGRKLLRLEVRNAETPIERKPPWIKTRAKMGPEYK
Mvan_3582|M.vanbaalenii_PYR-1 MTIAPRSDVAGHAAPEGRKLLRLEVRNAETPIERKPPWIKTRAKMGPEYK
TH_0219|M.thermoresistible__bu VTVTP----------EGRKLLRLEVRNAQTPIERKPPWIKTRATMGPEYR
MSMEG_4286|M.smegmatis_MC2_155 MTVVP----------EGRKLLRLEVRNAQTPIERKPPWIKTRAKMGPEYT
Mb2241|M.bovis_AF2122/97 MSVAAE----------GRRLLRLEVRNAQTPIERKPPWIKTRARIGPEYT
Rv2218|M.tuberculosis_H37Rv MSVAAE----------GRRLLRLEVRNAQTPIERKPPWIKTRARIGPEYT
MAV_2270|M.avium_104 MTVAPE----------GRKLLRLEVRNADTPIERKPPWIRVRARMGPEYT
MLBr_00858|M.leprae_Br4923 MTVAPE----------DCRLLRLEVRNAQTPIERKPPWIKTRARMGPEYT
MMAR_3286|M.marinum_M MNVAPEPGAAGAAAPQGRKLLRLEVRNAQTPIERKPPWIRTRARMGPEYT
MUL_1343|M.ulcerans_Agy99 MNVAPEPGAAGAAAPQGRKLLRLEVRNAQTPIERKPPWIRTRARMGPEYT
MAB_1942c|M.abscessus_ATCC_199 MTEP-------------RKLLRLEVRNAQTPIERKPDWIRTRAKMGPEYK
:. :*********:******* **:.** :****
Mflv_2932|M.gilvum_PYR-GCK ELKALVRREGLHTVCEEAGCPNIFECWEDREATFLIGGEQCTRRCDFCQI
Mvan_3582|M.vanbaalenii_PYR-1 ELKGLVRREGLHTVCEEAGCPNIFECWEDREATFLIGGEQCTRRCDFCQI
TH_0219|M.thermoresistible__bu DLKALVRREGLHTVCEEAGCPNIFECWEDREATFLIGGEQCTRRCDFCQI
MSMEG_4286|M.smegmatis_MC2_155 ELKGLVRREGLHTVCEEAGCPNIFECWEDREATFLIGGEQCTRRCDFCQI
Mb2241|M.bovis_AF2122/97 ELKNLVRREGLHTVCEEAGCPNIFECWEDREATFLIGGDQCTRRCDFCQI
Rv2218|M.tuberculosis_H37Rv ELKNLVRREGLHTVCEEAGCPNIFECWEDREATFLIGGDQCTRRCDFCQI
MAV_2270|M.avium_104 ELKSLVRREGLHTVCEEAGCPNIFECWEDREATFLIGGDQCTRRCDFCQI
MLBr_00858|M.leprae_Br4923 ALKNLVRRVALHTVCEEAGCPNIFECWEDREATFLIGGDQCTRRCDFCQI
MMAR_3286|M.marinum_M ELKNLVRREGLHTVCEEAGCPNIFECWEDREATFLIGGDQCTRRCDFCQI
MUL_1343|M.ulcerans_Agy99 ELKNLVRREGLHTVCEEAGCPNIFECWEDREATFLIGGDQCTRRCDFCQI
MAB_1942c|M.abscessus_ATCC_199 ELKSLVRSGGLHTVCEEAGCPNIFECWEDREATFLIGGEQCTRRCDFCQI
** *** .****************************:***********
Mflv_2932|M.gilvum_PYR-GCK DTGKPADLDRDEPRRVAESVQAMGLRYSTVTGVARDDLPDGGAWLYAETV
Mvan_3582|M.vanbaalenii_PYR-1 DTGKPAELDRDEPRRVAESVAAMGLRYSTVTGVARDDLPDGGAWLYSETV
TH_0219|M.thermoresistible__bu DTGKPAELDTDEPRRVAESVHAMGLRYSTVTGVARDDLPDGGAWLYAETV
MSMEG_4286|M.smegmatis_MC2_155 DTGKPADLDRDEPRRVAESVQAMGLRYSTVTGVARDDLPDGGAWLYAETV
Mb2241|M.bovis_AF2122/97 DTGKPAELDRDEPRRVADSVRTMGLRYATVTGVARDDLPDGGAWLYAATV
Rv2218|M.tuberculosis_H37Rv DTGKPAELDRDEPRRVADSVRTMGLRYATVTGVARDDLPDGGAWLYAATV
MAV_2270|M.avium_104 DTGKPAELDRDEPRRVADSVRTMGLRYATVTGVARDDLPDGGAWLYAETV
MLBr_00858|M.leprae_Br4923 DTGKPAALDRDEPRRVAESVQTMGLRYTTVTGVARDDLPDGGAWLYATTV
MMAR_3286|M.marinum_M DTGKPAPLDRDEPRRVAESVHTMGLRYATVTGVARDDLPDGGAWLYAETV
MUL_1343|M.ulcerans_Agy99 DTGKPAPLDRDEPRRVAESVHTMGLRYATVTGVARDDLPDGGAWLYAETV
MAB_1942c|M.abscessus_ATCC_199 DTGKPADLDRDEPRRVAESVHTMGLRYSTVTGVARDDLPDGGAWLYAETV
****** ** *******:** :*****:******************: **
Mflv_2932|M.gilvum_PYR-GCK RQIKALNPNTGVELLIPDFNADPDQLRAVFESRPEVLAHNVETVPRIFKR
Mvan_3582|M.vanbaalenii_PYR-1 RQIKALNPNTGVELLIPDFNADPAQLQQVFESRPEVLAHNVETVPRIFKR
TH_0219|M.thermoresistible__bu RAIKALNPNTGVELLIPDFNGDPELLEQVFETRPEVLAHNVETVPRIFKR
MSMEG_4286|M.smegmatis_MC2_155 RYIKRLNPNTGVELLIPDFNGNPEQLEEVFESRPEVLAHNVETVPRIFKR
Mb2241|M.bovis_AF2122/97 RAIKELNPSTGVELLIPDFNGEPTRLAEVFESGPEVLAHNVETVPRIFKR
Rv2218|M.tuberculosis_H37Rv RAIKELNPSTGVELLIPDFNGEPTRLAEVFESGPEVLAHNVETVPRIFKR
MAV_2270|M.avium_104 RAIKELNPSTGVELLIPDFNGRPDRLAEVFGSRPEVLAHNVETVPRIFKR
MLBr_00858|M.leprae_Br4923 RAIKELNPSTGVELLIPDFNGQPARLAEVFDSRPQVLAHNVETVPRIFKR
MMAR_3286|M.marinum_M RAIKELNPSTGVELLIPDFNGKADQLGEVFEARPEVLAHNVETVPRVFKR
MUL_1343|M.ulcerans_Agy99 RAIKELNPSTGVELLIPDFNGKADQLGEVFEARPEVLAHNVETVPRVFKR
MAB_1942c|M.abscessus_ATCC_199 RYIKDLNPATGVELLIPDFNAIPAQLDEVFASRPEVLAHNLETVPRVFRR
* ** *** ***********. . * ** : *:*****:*****:*:*
Mflv_2932|M.gilvum_PYR-GCK IRPGFRYERSLAVITAARDYGLVTKSNLILGMGETPEEVRAALHDLHDAG
Mvan_3582|M.vanbaalenii_PYR-1 IRPGFRYERSLSVLTMARDYGLVTKSNLILGMGETPEEVREALVDLHRAG
TH_0219|M.thermoresistible__bu IRPAFRYDRSLAVLTAARDYGLVTKSNLILGLGETPDEVRTALRDLHEAG
MSMEG_4286|M.smegmatis_MC2_155 IRPAFRYDRSLAVITAARNFGLVTKSNLILGMGETIDEVRTALRDLHDAG
Mb2241|M.bovis_AF2122/97 IRPAFTYRRSLGVLTAARDAGLVTKSNLILGLGETSDEVRTALGDLRDAG
Rv2218|M.tuberculosis_H37Rv IRPAFTYRRSLGVLTAARDAGLVTKSNLILGLGETSDEVRTALGDLRDAG
MAV_2270|M.avium_104 IRPAFTYRRSLDVLTAAREAGLVTKSNLILGLGETPDEVRTALADLRGAG
MLBr_00858|M.leprae_Br4923 IRPAFTYQRSLDVLTAAREAGLVTKSNLILGLGETADEVRTALADLRKTG
MMAR_3286|M.marinum_M IRPAFTYQRSLDVLTAARQFGLVTKSNLILGMGETPEEVRTALVDLHDAG
MUL_1343|M.ulcerans_Agy99 IRPAFTYQRSLDVLTAARQFGLVTKSNLILGMGETPEEVRTALVDLHDAG
MAB_1942c|M.abscessus_ATCC_199 IRPAFSYERSLSVITAAREAQLVTKSNLILGMGETNDEIREALVALHEAG
***.* * *** *:* **: **********:*** :*:* ** *: :*
Mflv_2932|M.gilvum_PYR-GCK CDIVTITQYLRPSPRHHPVERWVHPDEFVDHERYATEIGFAGVLAGPLVR
Mvan_3582|M.vanbaalenii_PYR-1 CDIVTITQYLRPSARHHPVERWVHPDEFVEHERYATGLGFAGVLAGPLVR
TH_0219|M.thermoresistible__bu CDIVTITQYLRPSPRHHPVERWVHPDEFVEYEQFARELGFAGVLAGPLVR
MSMEG_4286|M.smegmatis_MC2_155 CDIITITQYLRPSPRHHPVERWVHPDEFVELSAYAEGLGFAGVLAGPLVR
Mb2241|M.bovis_AF2122/97 CDIVTITQYLRPSARHHPVERWVKPEEFVQFARFAEGLGFAGVLAGPLVR
Rv2218|M.tuberculosis_H37Rv CDIVTITQYLRPSARHHPVERWVKPEEFVQFARFAEGLGFAGVLAGPLVR
MAV_2270|M.avium_104 CDIITITQYLRPSARHHPVERWVKPEEFVEFARHAEELGFSGVLAGPLVR
MLBr_00858|M.leprae_Br4923 CDIVTITQYLRPSMRHHPVERWVRPEEFVEYTQYAEGLGFSGVLGGPLVR
MMAR_3286|M.marinum_M CDIITITQYLRPSPRHHPVERWVRPEEFVEFAQYAEGLGFAGVLAGPLVR
MUL_1343|M.ulcerans_Agy99 CDIITITQYLRPSPRHHPVERWVRPEEFVEFAQYAEGLGFAGVLAGPLVR
MAB_1942c|M.abscessus_ATCC_199 CDIITITQYLRPSPRHHPVDRWVKPQEFVELSEFAEGLGFAGVMAGPLVR
***:********* *****:***:*:***: .* :**:**:.*****
Mflv_2932|M.gilvum_PYR-GCK SSYRAGKLYAQTVAKR-----SAVSLSNGEIA----------
Mvan_3582|M.vanbaalenii_PYR-1 SSYRAGKLYAQTIASRPTGAAPAAMPTSTSVS----------
TH_0219|M.thermoresistible__bu SSYRAGRLYAQVAQSR-----PPVS-----------------
MSMEG_4286|M.smegmatis_MC2_155 SSYRAGKLYAQAARVRFADQ-PPVS-----------------
Mb2241|M.bovis_AF2122/97 SSYRAGRLYEQARNSRALASR---------------------
Rv2218|M.tuberculosis_H37Rv SSYRAGRLYEQARNSRALASR---------------------
MAV_2270|M.avium_104 SSYRAGRLYRQAARARA-------------------------
MLBr_00858|M.leprae_Br4923 SSYRAGRLYEQAAGTRTVGTSVSR------------------
MMAR_3286|M.marinum_M SSYRAGRLYEQARSRNTSGASNSG------------------
MUL_1343|M.ulcerans_Agy99 SSYRAGRLYEQARSRNTSGASNSG------------------
MAB_1942c|M.abscessus_ATCC_199 SSYRAGRLYAQAMAHHGRPLPAALEHLATAGSARQEAINLIG
******:** *. .