For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGPNPTEPDANGSDLPDQTPGKHAVPEEPTQVSGTDPETGETEFYSQAYSAPEAEQYTAGPYVPADVALY DYDNYDPASELVEGPPPRWPWVVGVTAIIAAIALVASVALLVARDDQTSNLATPRPSTTTSSAPPVQDEI TTTTPPPVEPPPPPPPSEEAPPPPPPETVTITEPPPAPAPPPSSEEPAPPPATTTAPPATTTTPPAGPRQ VTYSVTGTKAPGDIITITYVDGNGNRRTLRNVYIPWTFTMTPISNSDVGSVEASSLFLVSRLNCSITASD GTVLSSNANNSAQTAC*
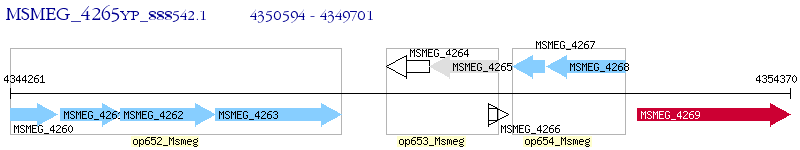
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4265 | - | - | 100% (297) | MmpS3 protein |
| M. smegmatis MC2 155 | MSMEG_1932 | - | 9e-30 | 34.64% (280) | MmpS3 protein |
| M. smegmatis MC2 155 | MSMEG_0638 | - | 5e-19 | 31.27% (259) | hypothetical protein MSMEG_0638 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2221c | mmpS3 | 8e-98 | 62.66% (308) | membrane protein |
| M. gilvum PYR-GCK | Mflv_2951 | - | 1e-104 | 64.55% (299) | hypothetical protein Mflv_2951 |
| M. tuberculosis H37Rv | Rv2198c | mmpS3 | 9e-98 | 62.66% (308) | membrane protein |
| M. leprae Br4923 | MLBr_00877 | mmpS3 | 4e-84 | 55.13% (312) | hypothetical protein MLBr_00877 |
| M. abscessus ATCC 19977 | MAB_1963 | - | 8e-84 | 56.38% (298) | MmpS family protein |
| M. marinum M | MMAR_3242 | mmpS3 | 3e-99 | 60.68% (323) | membrane protein MmpS3 |
| M. avium 104 | MAV_2293 | - | 2e-73 | 52.82% (301) | MmpS3 protein |
| M. thermoresistible (build 8) | TH_1385 | - | 8e-19 | 28.22% (287) | POSSIBLE CONSERVED PROLINE RICH MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_3555 | mmpS3 | 2e-99 | 60.68% (323) | membrane protein MmpS3 |
| M. vanbaalenii PYR-1 | Mvan_3562 | - | 1e-103 | 63.88% (299) | putative membrane protein MmpS3 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3242|M.marinum_M MSGP-NPPGWEPDEPD-PAGDSSAELEPFGDTDDE------PDDALAGGE
MUL_3555|M.ulcerans_Agy99 MSGP-NPPGWEPDEPD-PAGDSSAELEPFGDTGDE------PDDALAGGE
Mb2221c|M.bovis_AF2122/97 MSGP-NPPGREPDEPE--SEPVSDTGDERA----------------SGNH
Rv2198c|M.tuberculosis_H37Rv MSGP-NPPGREPDEPE--SEPVSDTGDERA----------------SGNH
MLBr_00877|M.leprae_Br4923 MSGP-NPPGRENEESD-SGNELSGELDPHN-------------GVESVDE
MAV_2293|M.avium_104 MSGS-RPPGWEPEEPDPAADPIDHQPDPGGPPADEFPGEDFPAGGDHEDA
Mflv_2951|M.gilvum_PYR-GCK MSGP-NPGE-----------------------------------------
Mvan_3562|M.vanbaalenii_PYR-1 MSGP-NPPEEPGGQPA----------------------------------
MSMEG_4265|M.smegmatis_MC2_155 MSGP-NPTEPDANGSD--------LPDQTP-----------------GKH
MAB_1963|M.abscessus_ATCC_1997 MSGTDNPGSGNPGEVP---------------------------------D
TH_1385|M.thermoresistible__bu MTESPRRGGTDPTEPM----------------------------------
*: . .
MMAR_3242|M.marinum_M LEPVSESGQIEPVDPAQESEAYSRAYSAPESEQFISGAYLPADLSLYDYD
MUL_3555|M.ulcerans_Agy99 LEPVSESGQIEPVDPAQESEAYSRAYSAPESEQFISGAYLPADLSLYDYD
Mb2221c|M.bovis_AF2122/97 LPPVAGGGDKLPSDQTGETDAYSRAYSAPESEHVTGGPYVPADLRLYDYD
Rv2198c|M.tuberculosis_H37Rv LPPVAGGGDKLPSDQTGETDAYSRAYSAPESEHVTGGPYVPADLRLYDYD
MLBr_00877|M.leprae_Br4923 LVPVPDSDLVTASDHTSETEVYSQAYSAPEAEHFTAVPYVPADLRLYDYD
MAV_2293|M.avium_104 VEPFDDSVEFDAADQTGQTDAYSRAYSAPESEHFTSGPYVPADLRLYDYD
Mflv_2951|M.gilvum_PYR-GCK ---PDNAA---DNPDTGQTDFYSRAYSAPESEQFVS-AYSAADIVPYDYD
Mvan_3562|M.vanbaalenii_PYR-1 ---PDDATRGVDNPDTGQTDFYSRAYSAPESEQFVSPPYVPADASLYDYD
MSMEG_4265|M.smegmatis_MC2_155 AVPEEPTQVSGTDPETGETEFYSQAYSAPEAEQYTAGPYVPADVALYDYD
MAB_1963|M.abscessus_ATCC_1997 LDGPNEEAAAPDSSGADSAAFHSQAYSAPESEQFTS-PYVP-------YD
TH_1385|M.thermoresistible__bu -YPGYSDPAYASQAYGPTYPASPQPPPTEQLPAYPPYGYDPYGTGQQGYE
.:. .: : * . *:
MMAR_3242|M.marinum_M GYDDSLDEEDEPSTPRWPWVVGVAAIMAAIALVASVSLLVT-RTD-STKL
MUL_3555|M.ulcerans_Agy99 GYDDSLDEEDEPSTPRWPWVVGVAAIMAAIALVASVSLLVT-RTD-STKL
Mb2221c|M.bovis_AF2122/97 DYEESSDLDDELAAPRWPWVVGVAAIIAAVALVVSVSLLVT-RPH-TSKL
Rv2198c|M.tuberculosis_H37Rv DYEESSDLDDELAAPRWPWVVGVAAIIAAVALVVSVSLLVT-RPH-TSKL
MLBr_00877|M.leprae_Br4923 ---ESSVYDEPGAAPRWPWVVGVAAILAAISLVVSVSLLFT-RTD-TSKL
MAV_2293|M.avium_104 DYDEPAELDEEHGTARWPWVVGVAAIVAAIALVVSVSLLFA-RTD-TTKL
Mflv_2951|M.gilvum_PYR-GCK SYDATEEPAEFAPPPRWPWVVGVVAIVAAVALVVSVSVLVT-RTD-TDNL
Mvan_3562|M.vanbaalenii_PYR-1 TYDADAD-AELAPPPRWPWVVGIVAIVAAIALVVSVSVLVT-RSD-TDNL
MSMEG_4265|M.smegmatis_MC2_155 NYDPASELVEGP-PPRWPWVVGVTAIIAAIALVASVALLVA-RDDQTSNL
MAB_1963|M.abscessus_ATCC_1997 EYETQLVDGEEPPPPRWPWVVGVAAIIAAITLVASVALLVAGRSDETNGA
TH_1385|M.thermoresistible__bu YPPAEPPEPEKPKLPGWLWVLAALAVLLVVGLVIALVITNSSQQD-TVFA
: . * **:. *:: .: ** :: : : : . :
MMAR_3242|M.marinum_M ANP---ATTSSVPPVQDEITTTKPPPPPPPPPPPPSTEIPTATE----TQ
MUL_3555|M.ulcerans_Agy99 ANP---ATTSSVPPVQDEITTTKPPPPPPPPPPPPSTEIPTATE----TQ
Mb2221c|M.bovis_AF2122/97 ATG---DTTSSAPPVQDEITTTKPAPPPPPPAPPPTTEIPTATE----TQ
Rv2198c|M.tuberculosis_H37Rv ATG---DTTSSAPPVQDEITTTKPAPPPPPPAPPPTTEIPTATE----TQ
MLBr_00877|M.leprae_Br4923 STP---TTGRSTPPVQDEITTVKP--------PPPSTETSTATE----TQ
MAV_2293|M.avium_104 ANPN--TTTASTPPLQDEITTTKP--------PPP---------------
Mflv_2951|M.gilvum_PYR-GCK ATPE--TTTRQAPPVQDEILTTTP--PPPPPPPPTTEPPPPPPP----PE
Mvan_3562|M.vanbaalenii_PYR-1 ATPQ--TTTRPAPPVQDEFTTTEP--PPPP----TTEPPPPPP------E
MSMEG_4265|M.smegmatis_MC2_155 ATPRPSTTTSSAPPVQDEITTTTPPPVEPPPPPPPSEEAPPPPP----PE
MAB_1963|M.abscessus_ATCC_1997 AKSSTVIPTPSTTPWNEIITTTTP--PPPPPPPPPTTEPPPPPPPPVVTE
TH_1385|M.thermoresistible__bu PGP---SATEPSFPMPTPPTTTRR---------APTTVLPPLPS------
. . * *. .
MMAR_3242|M.marinum_M TVTVTPPPPPPPPPETSAAPPPAATTPTTPAAAPPPTTTTPAGPRQVTYS
MUL_3555|M.ulcerans_Agy99 TVTVTPPPPPPPPPETSAAPPPAATTPTTPAAAPPPTTTTPAGPRQVTYS
Mb2221c|M.bovis_AF2122/97 TVTVTPPPPPPP----ATTTAPPPATTTTAAAPPPTT-TTPTGPRQVTYS
Rv2198c|M.tuberculosis_H37Rv TVTVTPPPPPPP----ATTTAPPPATTTTAAAPPPTT-TTPTGPRQVTYS
MLBr_00877|M.leprae_Br4923 TVTVTPLPPPSA----TSTAVPPSSVVPPPPTTPTTTVTTLTGPRQVTYS
MAV_2293|M.avium_104 --------------------------------------TTPTGPRQVTYS
Mflv_2951|M.gilvum_PYR-GCK TVTVTQPPPPPP-----AEAPPPPPPQTTEAPPPPPTTTTPAGPRQVTYS
Mvan_3562|M.vanbaalenii_PYR-1 TVTVTEPPPPPPE---TTAAPPPPPPATTEPPPPPPTTTTRAGPRQVTYS
MSMEG_4265|M.smegmatis_MC2_155 TVTITEPPPAPAP---PPSSEEPAPPPATTTAPPATTTTPPAGPRQVTYS
MAB_1963|M.abscessus_ATCC_1997 TVTVEPPPPPPPP-VTTHEAPPPVTTTTTPPPPPPPTTTTPAGPRQITYS
TH_1385|M.thermoresistible__bu -----------------------PGPTTGPTTGPPGATTTPGATETVVYE
*. ... :.*.
MMAR_3242|M.marinum_M VTGTKAPGDIISVTYVDASGRRRTQHNVYIPWSMTVTPISQSDVGSVEAS
MUL_3555|M.ulcerans_Agy99 VTGTKAPGDIISVTYVDASGRRRTQHNVYIPWSMTVTPISQSDVGSVEAS
Mb2221c|M.bovis_AF2122/97 VTGTKAPGDIISVTYVDAAGRRRTQHNVYIPWSMTVTPISQSDVGSVEAS
Rv2198c|M.tuberculosis_H37Rv VTGTKAPGDIISVTYVDAAGRRRTQHNVYIPWSMTVTPISQSDVGSVEAS
MLBr_00877|M.leprae_Br4923 VTGTKAPGDIISVTYVDASGRRRTQHNVYIPWSMTVTPISQSDVGSVEAF
MAV_2293|M.avium_104 VTGTKAPGDIISVTYVDASGRRRTQHNVYIPWSMTVTPISQSDVGSVEAS
Mflv_2951|M.gilvum_PYR-GCK VTGTKAPGDIITVTYIDASGRSRTQRNVYIPWSLTVTPISQSEVGSVQAS
Mvan_3562|M.vanbaalenii_PYR-1 VTGTKAPGDIITVTYIDASGRSRTQRNVYIPWSLTVTPISQSEVGSVQAS
MSMEG_4265|M.smegmatis_MC2_155 VTGTKAPGDIITITYVDGNGNRRTLRNVYIPWTFTMTPISNSDVGSVEAS
MAB_1963|M.abscessus_ATCC_1997 VTGSKAPLDRISITWTDGSGRTRVNPNVYIPWSITVTPISNSEIGSVSAS
TH_1385|M.thermoresistible__bu VAG---TGRAINITYIDTGKVLQTEFNVQLPWRKQVELPSP--AASSASV
*:* . *.:*: * :. ** :** : * .* :
MMAR_3242|M.marinum_M SLFRVSRLNCSITTSDGTVLSSNSNDAPQTSC---
MUL_3555|M.ulcerans_Agy99 SLFRVSRLNCSITTSDGTVLSSNSNDAPQTSC---
Mb2221c|M.bovis_AF2122/97 SLFRVSKLNCSITTSDGTVLSSNSNDGPQTSC---
Rv2198c|M.tuberculosis_H37Rv SLFRVSKLNCSITTSDGTVLSSNSNDGPQTSC---
MLBr_00877|M.leprae_Br4923 SLFRVSKLNCLITTSDGTVLSSNSNDAPQTSC---
MAV_2293|M.avium_104 SLFRVSRLNCSITTSDGTVLSSNSNDAPQTSC---
Mflv_2951|M.gilvum_PYR-GCK SLFLVSKLNCSITTSDGQVLSSNTGNAAQTSC---
Mvan_3562|M.vanbaalenii_PYR-1 SLFMVSRLNCSITTSDGTVLSSNTSNAAQTSC---
MSMEG_4265|M.smegmatis_MC2_155 SLFLVSRLNCSITASDGTVLSSNANNSAQTAC---
MAB_1963|M.abscessus_ATCC_1997 SFLRLSQLNCTITTSDGQVLSSNNNNSAQATC---
TH_1385|M.thermoresistible__bu SIINVGRDVTCTVTVDGALIQQRTGSGLTICVGAG
*:: :.: .: ** ::... ...