For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RFRQLWAAVVAVLLAVTMVTACGVTRGDESRGLHRLRMMVPNSPGGGYDLTARTAVKIMEDTDITGRIEV FNVIGAGGTVAMARLMNERGNNDLMMMMGLGVVGAVYTNGSSARASDATALAKMVEEQEGILVPGDSPFR TIDDLVAAWKADPSKVSIGGGSSPGGPDHLFPMETARAVGVDPRTVNYITYDGGGDLLTALLGKKIAAGT TGLGEFVDQIEAGQVRVLAVSGAERVPGVDAPTLTEAGIDMTFTNWRGVLAPPGISDDARDAMVAALTEL HDTDEWKEAMVKNGWSDAFTTGPEFEQFLKDQDERVSTTLTDLGLL*
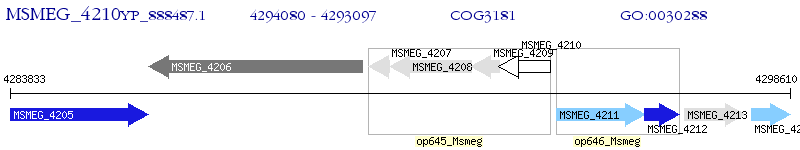
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4210 | - | - | 100% (327) | secreted protein |
| M. smegmatis MC2 155 | MSMEG_0122 | - | 3e-19 | 25.30% (332) | putative periplasmic solute-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3004 | - | 1e-146 | 77.78% (324) | hypothetical protein Mflv_3004 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3502 | - | 1e-149 | 80.75% (322) | hypothetical protein Mvan_3502 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3004|M.gilvum_PYR-GCK MIGDDMFA--RRRTIVGVVIAAVVTLLLCSCGVTRGEET-GLHRLRMMVP
Mvan_3502|M.vanbaalenii_PYR-1 MIGDDMFASSRRGTLIGLTIAVIAALLLTGCGVTRGEQT-GLHRLRMMVP
MSMEG_4210|M.smegmatis_MC2_155 MRFRQLWA--------AVVAVLLAVTMVTACGVTRGDESRGLHRLRMMVP
* :::* .:. . :.. :: .******::: **********
Mflv_3004|M.gilvum_PYR-GCK NSPGGGYDLTARTAVKIMEDDAITGRVEVFNVIGAGGTVAMARLMNERGN
Mvan_3502|M.vanbaalenii_PYR-1 NSPGGGYDLTARTAVKIMEDDEITGRVEVFNVIGAGGTVAMARLMNEQGN
MSMEG_4210|M.smegmatis_MC2_155 NSPGGGYDLTARTAVKIMEDTDITGRIEVFNVIGAGGTVAMARLMNERGN
******************** ****:********************:**
Mflv_3004|M.gilvum_PYR-GCK GDLMMMMGLGVVGAVFTNGSTARASDATALAKVVEEQEGILVPADSPFQT
Mvan_3502|M.vanbaalenii_PYR-1 GDLMMMMGLGVVGATFTNGSKARASDATALAKMVEEQEGILVPADSPFTT
MSMEG_4210|M.smegmatis_MC2_155 NDLMMMMGLGVVGAVYTNGSSARASDATALAKMVEEQEGILVPGDSPFRT
.*************.:****.***********:**********.**** *
Mflv_3004|M.gilvum_PYR-GCK VQDFVDAWKADPAKVTIGGGSNPGGPDHLFPMETAKAVGVDPTKVNFVSY
Mvan_3502|M.vanbaalenii_PYR-1 VQDFVAAWKADPAKVTIGGGSNPGGPDHLFPMETARAVGVDPTKVNFVSY
MSMEG_4210|M.smegmatis_MC2_155 IDDLVAAWKADPSKVSIGGGSSPGGPDHLFPMETARAVGVDPRTVNYITY
::*:* ******:**:*****.*************:****** .**:::*
Mflv_3004|M.gilvum_PYR-GCK DGGGDLLTALLGNKISAGTSGLGEYVDQIEAGQVRVLAVSGEERVEGVDA
Mvan_3502|M.vanbaalenii_PYR-1 DGGGDLLTALLGNKIAAGTSGLGEYVDQIESGQVRVLAVSGEERVEGVDA
MSMEG_4210|M.smegmatis_MC2_155 DGGGDLLTALLGKKIAAGTTGLGEFVDQIEAGQVRVLAVSGAERVPGVDA
************:**:***:****:*****:********** *** ****
Mflv_3004|M.gilvum_PYR-GCK PTLKEAGIDLTFTNWRGVLAPPGISEEDRQALVRILEELHATEAWKEALV
Mvan_3502|M.vanbaalenii_PYR-1 PTLTEAGIDLTFTNWRGVIAPPGISDQDRAAMVKVLEELHATEAWKEALV
MSMEG_4210|M.smegmatis_MC2_155 PTLTEAGIDMTFTNWRGVLAPPGISDDARDAMVAALTELHDTDEWKEAMV
***.*****:********:******:: * *:* * *** *: ****:*
Mflv_3004|M.gilvum_PYR-GCK TNGWSDAFMTGQPFEDFLAEQDQRVESTLTDLGLV
Mvan_3502|M.vanbaalenii_PYR-1 KNGWSDAFMTGEPFEEFLTEQDKRVETTLTDLGLV
MSMEG_4210|M.smegmatis_MC2_155 KNGWSDAFTTGPEFEQFLKDQDERVSTTLTDLGLL
.******* ** **:** :**:**.:*******: