For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAIPALRRTRLVLAAAAAAGLVLSGCGGVQNTTGGGSGGTYPSGTVEMYVGASAGGSSDLISRAVSKGLS DSLGASFPVINREGANGALAAAEVSKAKPDGSVIAIQNASLFTITPLAVSPGEVTDIDDFDVVYGISRDD YVLVTNPASGYKTMGDLEAATKPVRYGTTGVGTGAQLAAALLFKSADVPSQAVPFDGGAPALTALLGNQI DVAVLQVGEAIENIQSDKLIPLSVFGPERIDYLPEVPTAKEQGLDVEVTQYRFMTVPKGTPQEVRDRIVE GLEATFATDAYKKFNEQNSLTPMEQPGPEVLEQLKADSKRYADLTKEFGIDLRAS
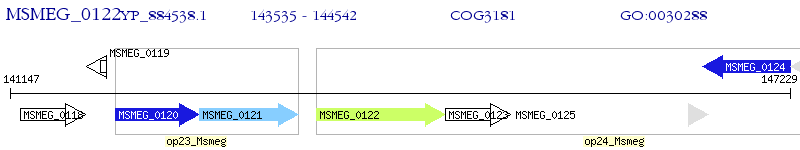
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0122 | - | - | 100% (335) | putative periplasmic solute-binding protein |
| M. smegmatis MC2 155 | MSMEG_4210 | - | 3e-19 | 25.30% (332) | secreted protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2741 | - | 5e-24 | 31.27% (323) | hypothetical protein Mflv_2741 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3502 | - | 3e-21 | 28.44% (334) | hypothetical protein Mvan_3502 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2741|M.gilvum_PYR-GCK -----MFRS-------ALGAVMAAALVLTGCQ---KSEEPVAAPPYPAGP
Mvan_3502|M.vanbaalenii_PYR-1 MIGDDMFASSRRGTLIGLTIAVIAALLLTGCG---VTRGEQTG----LHR
MSMEG_0122|M.smegmatis_MC2_155 -----MAIPALRRTRLVLAAAAAAGLVLSGCGGVQNTTGGGSGGTYPSGT
* . * . *.*:*:** : :.
Mflv_2741|M.gilvum_PYR-GCK VTMTAGANPGSGFDITIRAVVEALETEKLVTVPLPVQNKPGNIGADFLAS
Mvan_3502|M.vanbaalenii_PYR-1 LRMMVPNSPGGGYDLTARTAVKIMEDDEITGR-VEVFNVIGAGGTVAMAR
MSMEG_0122|M.smegmatis_MC2_155 VEMYVGASAGGSSDLISRAVSKGLSDS--LGASFPVINREGANGALAAAE
: * . ..*.. *: *:. : :. . . * * * *: *
Mflv_2741|M.gilvum_PYR-GCK MVEQYQGRDDQVSVTSLSMMMNQLRGLSP---YGYDDVTMIARLITEYYV
Mvan_3502|M.vanbaalenii_PYR-1 LMNE-QGNGDLMMMMGLGVVGATFTNGSK---ARASDATALAKMVEEQEG
MSMEG_0122|M.smegmatis_MC2_155 VSKA-KPDGSVIAIQNASLFTITPLAVSPGEVTDIDDFDVVYGISRDDYV
: : : .. : : . .:. * .* : : :
Mflv_2741|M.gilvum_PYR-GCK VVTDAQSPYRTLADVMTAIRSDPGRVQIAAATDDQAPFDLL----VAAAG
Mvan_3502|M.vanbaalenii_PYR-1 ILVPADSPFTTVQDFVAAWKADPAKVTIGGGSNPGGPDHLFPMETARAVG
MSMEG_0122|M.smegmatis_MC2_155 LVTNPASGYKTMGDLEAATKPVRYGTTGVGTGAQLAAALLFK--------
::. . * : *: *. :* :. . . .. *:
Mflv_2741|M.gilvum_PYR-GCK GDPAALDYVAFEGGGDQTEALLNSGISVAVGGVSEFVDLVRSGELRALGV
Mvan_3502|M.vanbaalenii_PYR-1 VDPTKVNFVSYDGGGDLLTALLGNKIAAGTSGLGEYVDQIESGQVRVLAV
MSMEG_0122|M.smegmatis_MC2_155 SADVPSQAVPFDGGAPALTALLGNQIDVAVLQVGEAIENIQSDKLIPLSV
. : *.::**. ***.. * ... :.* :: :.*.:: *.*
Mflv_2741|M.gilvum_PYR-GCK LSEERLPGL-DVPTAREQNLDVTLSNWRGLYGPPGMPAGAVSYWQEVLAE
Mvan_3502|M.vanbaalenii_PYR-1 SGEERVEGV-DAPTLTEAGIDLTFTNWRGVIAPPGISDQDRAAMVKVLEE
MSMEG_0122|M.smegmatis_MC2_155 FGPERIDYLPEVPTAKEQGLDVEVTQYRFMTVPKGTPQEVRDRIVEGLEA
. **: : :.** * .:*: .:::* : * * . : *
Mflv_2741|M.gilvum_PYR-GCK MVNTPTWNQIAERNQFTTTFMVGDELQTFLADTQADLAAALAQA------
Mvan_3502|M.vanbaalenii_PYR-1 LHATEAWKEALVKNGWSDAFMTGEPFEEFLTEQDKRVETTLTDLGLV---
MSMEG_0122|M.smegmatis_MC2_155 TFATDAYKKFNEQNSLTPMEQPGPEVLEQLKADSKRYADLTKEFGIDLRA
* :::: :* : * . * . :
Mflv_2741|M.gilvum_PYR-GCK -
Mvan_3502|M.vanbaalenii_PYR-1 -
MSMEG_0122|M.smegmatis_MC2_155 S