For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MENFDWLMQGFAEAATPMNLLYAVIGVLLGTAVGVLPGIGPAMTVALLLPVTYNVSPSAAFIMFAGIFYG GMYGGSTTSILLNTPGESSSVITAIEGNKMAKAGRAAQALATAAIGSFVAGAIGTALLAAFAPPISRFAV TLGAPSYLAIMLFALVAVTAVLGASKLRGAISLFLGLAIGCVGIDFLTGQPRATFGLPLLSDGIDIVVIA VAIFALGEALWVAAHLRRRPAEVIPVGRPWMDRADFGRSWKPWLRGTAFGFPFGALPAGGAELPTFLSYI TEKRLSKHKDEFGNGAIEGVAGPEAANNASAAGTLVPMLSLGLPTNATAAVMLTAFVSYGIQPGPTLFEK EPLLIWTLIASLFIGNFLLLVLNLPLAPLWARLLRTPRPYLYAGILFFATLGALAVNVQALDLALLLVFG LLGLMMRRFGLPVLPLIIGVILGPRIERQLRQSLQLGGGDWSSLFTEPVAIVVYVLMAILLLMPLVLKLM HRSEETLLIVEDDADQKEKAAQS*
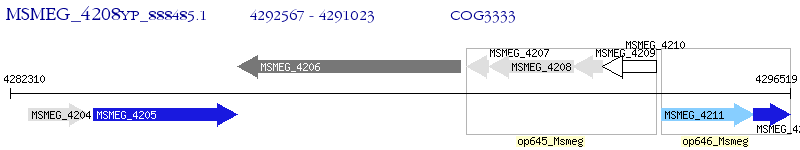
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4208 | - | - | 100% (514) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_0125 | - | e-112 | 41.32% (484) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_2051 | nuoM | 9e-05 | 25.61% (164) | NADH dehydrogenase subunit M |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3181 | nuoM | 1e-05 | 28.14% (167) | NADH dehydrogenase subunit M |
| M. gilvum PYR-GCK | Mflv_3006 | - | 0.0 | 91.42% (513) | hypothetical protein Mflv_3006 |
| M. tuberculosis H37Rv | Rv3157 | nuoM | 2e-06 | 28.74% (167) | NADH dehydrogenase subunit M |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2146 | - | 6e-05 | 26.47% (170) | NADH dehydrogenase subunit M |
| M. marinum M | MMAR_1471 | nuoM | 1e-06 | 29.34% (167) | NADH dehydrogenase I (chain M) NuoM (NADH- ubiquinone |
| M. avium 104 | MAV_4045 | - | 7e-07 | 29.17% (168) | NADH dehydrogenase subunit M |
| M. thermoresistible (build 8) | TH_1091 | nuoM | 2e-05 | 26.67% (165) | PROBABLE NADH DEHYDROGENASE I (CHAIN M) NUOK |
| M. ulcerans Agy99 | MUL_2471 | nuoM | 5e-07 | 29.34% (167) | NADH dehydrogenase subunit M |
| M. vanbaalenii PYR-1 | Mvan_3500 | - | 0.0 | 91.21% (512) | hypothetical protein Mvan_3500 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3181|M.bovis_AF2122/97 -MN-NVPWLSVLWLVPLAGAVLIILLPPGRRRLAKWAGMVVS-VLTLAVS
Rv3157|M.tuberculosis_H37Rv -MN-NVPWLSVLWLVPLAGAVLIILLPPGRRRLAKWAGMVVS-VLTLAVS
MMAR_1471|M.marinum_M -MNSHVPWLTLLWLVPLAASVLIILLPPGLRQLAKWTGLIAS-VVTLAVS
MUL_2471|M.ulcerans_Agy99 MVNSHVPWLTLLWLVPLAASVLTILLPPGLRQLAKWTGLIAS-VVTLAMS
MAV_4045|M.avium_104 -MN-QLPWLSILWLVPLAGSVLIILMPPGLRQLAKWTGLVFG-VLTLAVA
TH_1091|M.thermoresistible__bu -VVTGFPWLTALWATPIVGAAALVLLPAARRGLAKWFALAVS-LLVLGIT
MAB_2146|M.abscessus_ATCC_1997 --MSAVGAVTALWLIPLVGAAAILVLPAHQRTLAKYTALGTS-LLVLIVA
Mflv_3006|M.gilvum_PYR-GCK --MENFDWLLQGFSEAATPTNLMYAVIGVLLGTAVGVLPGIGPAMTVALL
Mvan_3500|M.vanbaalenii_PYR-1 --MENFDWLLQGFAEAATPTNLLYAVIGVLLGTAVGVLPGIGPAMTVALL
MSMEG_4208|M.smegmatis_MC2_155 -MMENFDWLMQGFAEAATPMNLLYAVIGVLLGTAVGVLPGIGPAMTVALL
. : : . . : * . :.: :
Mb3181|M.bovis_AF2122/97 IVVAAEFKPSAEPYQFVEKHSWIPAFGAGYTLGVDGIAVVLVLLTTVLIP
Rv3157|M.tuberculosis_H37Rv IVVAAEFKPSAEPYQFVEKHSWIPAFGAGYTLGVDGIAVVLVLLTTVLIP
MMAR_1471|M.marinum_M VVVATGFKPSGEHYQFVENRSWIPAFGASYTLGVDGIAVVLVLLTTVLIP
MUL_2471|M.ulcerans_Agy99 VVVATGFKPSGEHYQFVENRSWIPAFGASYTLGVDGIAVVLVLLTTVLIP
MAV_4045|M.avium_104 IVVTAGFKTGGAPYQFVEKHQWIPAFGAGYTLGVDGIAVVLVLLTAVLIP
TH_1091|M.thermoresistible__bu AVLTAGFDPGGERYQFVESHRWIPSFGTGYILGIDGIALALVLLTAVLVP
MAB_2146|M.abscessus_ATCC_1997 VGVAIAFDPVGPQYQFVESVSWIPAFGMKYAVGLDGIGLALVLLTTGLLP
Mflv_3006|M.gilvum_PYR-GCK LPITYNVSPSAAFIMFAG-----IFYGGMYGGSTTSILLNTPGESSSVIT
Mvan_3500|M.vanbaalenii_PYR-1 LPITYNVSPSAAFIMFAG-----IFYGGMYGGSTTSILLNTPGESSSVIT
MSMEG_4208|M.smegmatis_MC2_155 LPVTYNVSPSAAFIMFAG-----IFYGGMYGGSTTSILLNTPGESSSVIT
:: ... . *. :* * . .* : :: ::.
Mb3181|M.bovis_AF2122/97 LLLVAGWNDATDADDLSPASGRYPQRPAPPRLRSSGGERTR-GVHAYVAL
Rv3157|M.tuberculosis_H37Rv LLLVAGWNDATDADDLSPASGRYPQRPAPPRLRSSGGERTR-GVHAYVAL
MMAR_1471|M.marinum_M LLQIAGWND--------------------------GGERTR-GVHAYVAL
MUL_2471|M.ulcerans_Agy99 LLQIAGWND--------------------------GGERTR-GVHAYVAL
MAV_4045|M.avium_104 VLQVAGWND--------------------------GGDRQR-GVHAYVAL
TH_1091|M.thermoresistible__bu LLIVAGWNDADQ----------------------RPGLAGR-AVHLYLAL
MAB_2146|M.abscessus_ATCC_1997 VLLLAGWNDG----------------------AEAPGYGHRPLAQRYMAL
Mflv_3006|M.gilvum_PYR-GCK ALEGNKMAKA-------------------------GRAAQALATAAIGSF
Mvan_3500|M.vanbaalenii_PYR-1 AIEGNKMAKA-------------------------GRAAQALATAAIGSF
MSMEG_4208|M.smegmatis_MC2_155 AIEGNKMAKA-------------------------GRAAQALATAAIGSF
: . . ::
Mb3181|M.bovis_AF2122/97 TLAIESMVLMSVIALDVLLFYVFFEAMLIPMYFLIGGFGQ----GAGRSR
Rv3157|M.tuberculosis_H37Rv TLAIESMVLMSVIALDVLLFYVFFEAMLIPMYFLIGGFGQ----GAGRSR
MMAR_1471|M.marinum_M TLAIESMVLISVVALDVLLFYVFFEAMLIPMYFLIGGFGQ----GRARSG
MUL_2471|M.ulcerans_Agy99 TLAIESMVLISVVALDVLLFYVFFEAMLIPMYFLIGGFGQ----GRARSG
MAV_4045|M.avium_104 TLAIESMVLVSVIALDVLLFYVFFEAMLIPMYFLIGGFGA----GSGRSR
TH_1091|M.thermoresistible__bu TLAVEGMVLMSLIALDVLLFYVFFEAMLIPMYFLIGGFG-----GAHRGR
MAB_2146|M.abscessus_ATCC_1997 MLIVEAMVLLSFSALDVLLFYIFFEAMLIPMYFLIGGFGSTHSDVKQRSR
Mflv_3006|M.gilvum_PYR-GCK VAGAIGTTLLAAFAPAISRFAVTLGAPSYLAIMLFALVAVTAVLGSSKMR
Mvan_3500|M.vanbaalenii_PYR-1 VAGSIGTALLAAFAPMISRFAVTLGAPSYLAIMLFALVAVTAVLGSSKMR
MSMEG_4208|M.smegmatis_MC2_155 VAGAIGTALLAAFAPPISRFAVTLGAPSYLAIMLFALVAVTAVLGASKLR
. .*:: * : * : : * :*:. .. :
Mb3181|M.bovis_AF2122/97 AAVKFLLYNLFGGLIMLAAVIGLYVVTAQ-------YDSGTFDFREIVAG
Rv3157|M.tuberculosis_H37Rv AAVKFLLYNLFGGLIMLAAVIGLYVVTAQ-------YDSGTFDFREIVAG
MMAR_1471|M.marinum_M AAVKFLLYNLFGGLIMLAAVIGLYVVTAQ-------NGSGTFDFRDIVAG
MUL_2471|M.ulcerans_Agy99 AAVKFLLYNLFGGLIMLAAVIGLYVVTAQ-------NGSGTFDFRDIVAG
MAV_4045|M.avium_104 AAVKFLLYNLFGGLIMLAAVIGLYVVTSQ-------HGAGTFDFRDIVAG
TH_1091|M.thermoresistible__bu AAVKFLLYNLFGGLIMLAAVIGLYVVTAGSDALAGPAGGGTFDFRAIVAA
MAB_2146|M.abscessus_ATCC_1997 AAVKFLMYNLFGGLVMLAAVIGLYVVTAQSPAG---EHGGTFDLRDITEM
Mflv_3006|M.gilvum_PYR-GCK GVISLLLG-LAIGIVGIDSLTGQPRATFG-------LPLLSDGIDIVVIA
Mvan_3500|M.vanbaalenii_PYR-1 GLISLLLG-LAIGVVGIDSLTGQPRATFG-------IPLLSDGIDIVVIA
MSMEG_4208|M.smegmatis_MC2_155 GAISLFLG-LAIGCVGIDFLTGQPRATFG-------LPLLSDGIDIVVIA
. :.::: * * : : : * .* : .: :.
Mb3181|M.bovis_AF2122/97 VAAGRYGADPAVFKALFLGFMFAFAIKAPLWPFHRWLPDAAVESTPATAV
Rv3157|M.tuberculosis_H37Rv VAAGRYGADPAVFKALFLGFMFAFAIKAPLWPFHRWLPDAAVESTPATAV
MMAR_1471|M.marinum_M VQGGHYGEHPAVFKALFLGFMFAFAIKAPLWPFHRWLPDAAVESTPATAV
MUL_2471|M.ulcerans_Agy99 VQGGHYGEHPAVFKALFLGFMFAFAIKAPLWPFHRWLPDAAVESTPATAV
MAV_4045|M.avium_104 VASGRYGADPAVSKALFLGFMFAFAIKAPLWPFHRWLPDAAVEATPATAV
TH_1091|M.thermoresistible__bu VSAGDLMIDPAVLNLLFLGFMLAFAIKAPLWPFHRWLPDAAVEATPATAV
MAB_2146|M.abscessus_ATCC_1997 VATGTLGIDPGLAKALFLGFMFAFAVKAPLWPLHTWLPDAAVHATPSSAV
Mflv_3006|M.gilvum_PYR-GCK VAVFALGEALWVAAHLRRRPAEVIPVGRPWMGRDDWKRSWKPWLRGTAYG
Mvan_3500|M.vanbaalenii_PYR-1 VAVFAVGEALWVAAHLRRRPVDVIPVGRPWMSKQDWGRSWKPWLRGTAFG
MSMEG_4208|M.smegmatis_MC2_155 VAIFALGEALWVAAHLRRRPAEVIPVGRPWMDRADFGRSWKPWLRGTAFG
* : * .:.: * : . ::
Mb3181|M.bovis_AF2122/97 LMMAVMDKVGTFGMLRYCLQLFPDPSTYFRPLIVTLAIIGVIYGAIVAIG
Rv3157|M.tuberculosis_H37Rv LMMAVMDKVGTFGMLRYCLQLFPDPSTYFRPLIVTLAIIGVIYGAIVAIG
MMAR_1471|M.marinum_M LMMAVMDKVGTFGMLRYCLQLFPDASTYFRPLIVTLAIIGVIYGAVVAIG
MUL_2471|M.ulcerans_Agy99 LMMAVMDKVGTFGMLRYCLQLFPDASTYFRPLIVTLAIIGVIYGAVVAIG
MAV_4045|M.avium_104 LMMAVMDKVGTFGMLRYCLQLFPDAATYFRPLIVVLAVIGVIYGAIVAIG
TH_1091|M.thermoresistible__bu LMMAVMDKVGTFGMLRYSLQLFPDAATLFRPLVVALAVIGIVYGAILAIG
MAB_2146|M.abscessus_ATCC_1997 LMMAVVDKVGTFAMLRYCVQLFPDASRYFTPVIVTLAVIGIVYGAVVAIG
Mflv_3006|M.gilvum_PYR-GCK FPFGALPAGG--AELPTFLSYITEKRLTKHPEEFGKGAIEGVAGPEAANN
Mvan_3500|M.vanbaalenii_PYR-1 FPFGALPAGG--AELPTFLSYITEKKLSKHPEEFGKGAIEGVAGPEAANN
MSMEG_4208|M.smegmatis_MC2_155 FPFGALPAGG--AELPTFLSYITEKRLSKHKDEFGNGAIEGVAGPEAANN
: :..: * . * :. :.: . . * : *. * .
Mb3181|M.bovis_AF2122/97 QTDMMRLIAYTSISHFGFIIAGIFVMTTQG---QSGSTLYMLN--HGLST
Rv3157|M.tuberculosis_H37Rv QTDMMRLIAYTSISHFGFIIAGIFVMTTQG---QSGSTLYMLN--HGLST
MMAR_1471|M.marinum_M QTDIMRLIAYTSISHFGFIIAGIFVMTTQG---QSGSTLYMLN--HGLST
MUL_2471|M.ulcerans_Agy99 QTDIMRLIAYTSISHFGFIIAGIFVMTTQG---QSGSTLYMLN--HGLST
MAV_4045|M.avium_104 QADVMRLIAYTSISHFGFIILGIFVMTSQG---QSGSTLYMLN--HGLST
TH_1091|M.thermoresistible__bu QTDVMRLIAYTSISHFGFIILGIFVMTSQG---QSGSTLYMIN--HGLST
MAB_2146|M.abscessus_ATCC_1997 QTDVMRLIAYTSISHFGFIILGIFAMTSQG---QSGSTLYMVN--HGIST
Mflv_3006|M.gilvum_PYR-GCK ASAAGTLVPMLSLGLPTNATAAVMLTAFVSYGIQPGPTLFDKEPLLIWTL
Mvan_3500|M.vanbaalenii_PYR-1 ASAAGTLVPMLSLGLPTNATAAVILTAFVSYGIQPGPTLFDKEPLLIWTL
MSMEG_4208|M.smegmatis_MC2_155 ASAAGTLVPMLSLGLPTNATAAVMLTAFVSYGIQPGPTLFEKEPLLIWTL
: *:. *:. .:: : . *.*.**: : :
Mb3181|M.bovis_AF2122/97 AAVFLIAGFLIARRD-SRSIADYGGVQKVAP--ILAGTFMVSAMATVSLP
Rv3157|M.tuberculosis_H37Rv AAVFLIAGFLIARRG-SRSIADYGGVQKVAP--ILAGTFMVSAMATVSLP
MMAR_1471|M.marinum_M AAVFLIAGFLISRHG-SRAIADYGGVQKVAP--VLAGTFLVSAMATLSLP
MUL_2471|M.ulcerans_Agy99 AAVFLIAGFLISRHG-SRAIADYGGVQKVAP--VLAGTFLVSAMATLSLP
MAV_4045|M.avium_104 AAVFLIAGFLISRHGGSRAIADYGGVQKVAP--VLAGTFLVSAMATLSLP
TH_1091|M.thermoresistible__bu AALFLIAGFLVTRHG-TRLITDYGGVQKVAP--VLAGTFLVAGLATLSLP
MAB_2146|M.abscessus_ATCC_1997 AALMLVAGFLVTRKG-SRLIASYGGVQKVAP--VLAGSFLVAGLATLSLP
Mflv_3006|M.gilvum_PYR-GCK IASLFIGNFLLLALN-LPLAPLWAKLLRTPRPYLYAGILFFATLGAFAVN
Mvan_3500|M.vanbaalenii_PYR-1 IASLFIGNFLLLVLN-LPLAPLWARLLRTPRPYLYAGILFFATLGAFAVN
MSMEG_4208|M.smegmatis_MC2_155 IASLFIGNFLLLVLN-LPLAPLWARLLRTPRPYLYAGILFFATLGALAVN
* :::..**: . . :. : :.. : ** ::.: :.:.::
Mb3181|M.bovis_AF2122/97 GLAPFISEFLVLLGTFSRYWLAAAFGVTALVLSAVYMLWLYQRVMTGPIA
Rv3157|M.tuberculosis_H37Rv GLAPFISEFLVLLGTFSRYWLAAAFGVTALVLSAVYMLWLYQRVMTGPVA
MMAR_1471|M.marinum_M GLAPFISEFLVLLGTFNRYWLAAAFGVTALVLSAIYMLWLYQRVMTGPLA
MUL_2471|M.ulcerans_Agy99 GLAPFISEFLVLLGTFNRYWLAAALGVTALVLSAIYMLWLYQRVMTGPLA
MAV_4045|M.avium_104 GLAPFVSEFLVLLGTFNRYWLAAAFGVTALVLSAVYMLWLYQRMMTGPVA
TH_1091|M.thermoresistible__bu GLAPFISEFLVLIGTFTRYPVLAVLAATALVLSAIYILWTYQRMMAGPVR
MAB_2146|M.abscessus_ATCC_1997 GLAPFISEFLVLVGTFTRYPVAAACAVVALVLAAIYVLWMYQRMMTGPLA
Mflv_3006|M.gilvum_PYR-GCK -LQPLDLVLLLIFGLMGLMMRRFGLPVLPLIIGVILGPRIERQLRQSLQL
Mvan_3500|M.vanbaalenii_PYR-1 -LQPLDLVLLLIFGLMGLMMRRFGLPVLPLIIGVILGPRIERQLRQSLQL
MSMEG_4208|M.smegmatis_MC2_155 -VQALDLALLLVFGLLGLMMRRFGLPVLPLIIGVILGPRIERQLRQSLQL
: .: :*::.* : . .*::..: ::: .
Mb3181|M.bovis_AF2122/97 EGNERIGDLVGREMIVVAPLIALLLVLGVYPKPVLDIINPAVENTMTTIG
Rv3157|M.tuberculosis_H37Rv EGNERIGDLVGREMIVVAPLIALLLVLGVYPKPVLDIINPAVENTMTTIG
MMAR_1471|M.marinum_M KGNERIGDLVPREMIVVAPLIAMLLILGVYPKPVLDVINPAVEHTMSTIG
MUL_2471|M.ulcerans_Agy99 KGNERIGDLVPREMIVVAPLIAMLLILGVYPKPVLDVINPAVEHTMSTIG
MAV_4045|M.avium_104 GGNEKIGDLRARELVVVAPLIALLLVLGIYPKPVLDIINPAVENTMTTIG
TH_1091|M.thermoresistible__bu EDNRSIRDLIPRELVVVAPLIALLVGLGVYPKPALDIIDPAVGHTLTSIG
MAB_2146|M.abscessus_ATCC_1997 PGNETIGDLKRRELTVVAPLVALLLVLGIYPKPLLDIVNPAVEQTLRTVN
Mflv_3006|M.gilvum_PYR-GCK GGGEWGSLFTEPVAIVTYVLMALLLVA----PLVLRLMHRSEESLLIVED
Mvan_3500|M.vanbaalenii_PYR-1 GGGEWGSLFTEPVAIITYVLMILLLAA----PLVLRLMHRSEETLLVVED
MSMEG_4208|M.smegmatis_MC2_155 GGGDWSSLFTEPVAIVVYVLMAILLLM----PLVLKLMHRSEETLLIVED
.. : :. *: :*: * ::. : : .
Mb3181|M.bovis_AF2122/97 QHDPAPSVAHPVPAVGASRTAEGPHP
Rv3157|M.tuberculosis_H37Rv QHDPAPSVAHPVPAVGASRTAEGPHP
MMAR_1471|M.marinum_M QHDPAPKVPHPVSGVSAPRTAEGPHE
MUL_2471|M.ulcerans_Agy99 QHDPAPKVPHPVSGVSAPRTAEGPHE
MAV_4045|M.avium_104 QHDPAPSVP-----MGTPRTAEGRR-
TH_1091|M.thermoresistible__bu EHDPRPTIT-----AGDDPVPAGKGR
MAB_2146|M.abscessus_ATCC_1997 EKDPPPTVAD-----IALRHGEGEQR
Mflv_3006|M.gilvum_PYR-GCK DKDQREKASQS---------------
Mvan_3500|M.vanbaalenii_PYR-1 DRDQKEKAGKV---------------
MSMEG_4208|M.smegmatis_MC2_155 DADQKEKAAQS---------------
: * .