For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVNSHVPWLTLLWLVPLAASVLTILLPPGLRQLAKWTGLIASVVTLAMSVVVATGFKPSGEHYQFVENRS WIPAFGASYTLGVDGIAVVLVLLTTVLIPLLQIAGWNDGGERTRGVHAYVALTLAIESMVLISVVALDVL LFYVFFEAMLIPMYFLIGGFGQGRARSGAAVKFLLYNLFGGLIMLAAVIGLYVVTAQNGSGTFDFRDIVA GVQGGHYGEHPAVFKALFLGFMFAFAIKAPLWPFHRWLPDAAVESTPATAVLMMAVMDKVGTFGMLRYCL QLFPDASTYFRPLIVTLAIIGVIYGAVVAIGQTDIMRLIAYTSISHFGFIIAGIFVMTTQGQSGSTLYML NHGLSTAAVFLIAGFLISRHGSRAIADYGGVQKVAPVLAGTFLVSAMATLSLPGLAPFISEFLVLLGTFN RYWLAAALGVTALVLSAIYMLWLYQRVMTGPLAKGNERIGDLVPREMIVVAPLIAMLLILGVYPKPVLDV INPAVEHTMSTIGQHDPAPKVPHPVSGVSAPRTAEGPHE
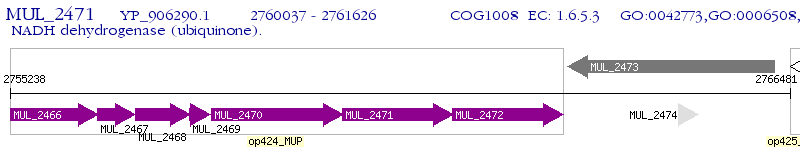
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2471 | nuoM | - | 100% (529) | NADH dehydrogenase subunit M |
| M. ulcerans Agy99 | MUL_2472 | nuoN | 6e-29 | 24.41% (381) | NADH dehydrogenase subunit N |
| M. ulcerans Agy99 | MUL_2470 | nuoL | 6e-26 | 25.43% (468) | NADH dehydrogenase subunit L |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3181 | nuoM | 0.0 | 83.48% (551) | NADH dehydrogenase subunit M |
| M. gilvum PYR-GCK | Mflv_4493 | - | 0.0 | 69.76% (506) | NADH dehydrogenase subunit M |
| M. tuberculosis H37Rv | Rv3157 | nuoM | 0.0 | 83.67% (551) | NADH dehydrogenase subunit M |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2146 | - | 0.0 | 67.56% (521) | NADH dehydrogenase subunit M |
| M. marinum M | MMAR_1471 | nuoM | 0.0 | 99.43% (528) | NADH dehydrogenase I (chain M) NuoM (NADH- ubiquinone |
| M. avium 104 | MAV_4045 | - | 0.0 | 84.54% (524) | NADH dehydrogenase subunit M |
| M. smegmatis MC2 155 | MSMEG_2051 | nuoM | 0.0 | 71.67% (526) | NADH dehydrogenase subunit M |
| M. thermoresistible (build 8) | TH_1091 | nuoM | 0.0 | 70.93% (516) | PROBABLE NADH DEHYDROGENASE I (CHAIN M) NUOK |
| M. vanbaalenii PYR-1 | Mvan_1873 | - | 0.0 | 70.35% (516) | NADH dehydrogenase subunit M |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2471|M.ulcerans_Agy99 MVNSHVPWLTLLWLVPLAASVLTILLPPGLRQLAKWTGLIASVVTLAMSV
MMAR_1471|M.marinum_M -MNSHVPWLTLLWLVPLAASVLIILLPPGLRQLAKWTGLIASVVTLAVSV
Mb3181|M.bovis_AF2122/97 -MN-NVPWLSVLWLVPLAGAVLIILLPPGRRRLAKWAGMVVSVLTLAVSI
Rv3157|M.tuberculosis_H37Rv -MN-NVPWLSVLWLVPLAGAVLIILLPPGRRRLAKWAGMVVSVLTLAVSI
MAV_4045|M.avium_104 -MN-QLPWLSILWLVPLAGSVLIILMPPGLRQLAKWTGLVFGVLTLAVAI
Mflv_4493|M.gilvum_PYR-GCK ----------------MVGAVAVLLLPAAARHLAKWTALVVSLVVLAVTG
Mvan_1873|M.vanbaalenii_PYR-1 -MVTSFPVLTALWAVPMVGAVVVMLLPAAARGLAKWVALSVSLIVLGVAA
MSMEG_2051|M.smegmatis_MC2_155 -MVSTFPWLTVLWAVPVVGAAVVILLPAAQQVLAKWLALAVSVLTLAVTA
TH_1091|M.thermoresistible__bu -VVTGFPWLTALWATPIVGAAALVLLPAARRGLAKWFALAVSLLVLGITA
MAB_2146|M.abscessus_ATCC_1997 --MSAVGAVTALWLIPLVGAAAILVLPAHQRTLAKYTALGTSLLVLIVAV
:..:. :::*. : ***: .: .::.* ::
MUL_2471|M.ulcerans_Agy99 VVATGFKPSGEHYQFVENRSWIPAFGASYTLGVDGIAVVLVLLTTVLIPL
MMAR_1471|M.marinum_M VVATGFKPSGEHYQFVENRSWIPAFGASYTLGVDGIAVVLVLLTTVLIPL
Mb3181|M.bovis_AF2122/97 VVAAEFKPSAEPYQFVEKHSWIPAFGAGYTLGVDGIAVVLVLLTTVLIPL
Rv3157|M.tuberculosis_H37Rv VVAAEFKPSAEPYQFVEKHSWIPAFGAGYTLGVDGIAVVLVLLTTVLIPL
MAV_4045|M.avium_104 VVTAGFKTGGAPYQFVEKHQWIPAFGAGYTLGVDGIAVVLVLLTAVLIPV
Mflv_4493|M.gilvum_PYR-GCK VIAVGFDPAGDQFQFVENYRWIPSFGTGYILGIDGIALALVVLTAVLLPL
Mvan_1873|M.vanbaalenii_PYR-1 VIAVGFDPAGDQFQFVENYRWIPSFGTGYILGVDGIALALVVLTAVLLPL
MSMEG_2051|M.smegmatis_MC2_155 VVAIGFDPAAAQYQFVESHRWIPSFGTGYILGVDGIALALVVLTAVLVPL
TH_1091|M.thermoresistible__bu VLTAGFDPGGERYQFVESHRWIPSFGTGYILGIDGIALALVLLTAVLVPL
MAB_2146|M.abscessus_ATCC_1997 GVAIAFDPVGPQYQFVESVSWIPAFGMKYAVGLDGIGLALVLLTTGLLPV
:: *.. . :****. ***:** * :*:***.:.**:**: *:*:
MUL_2471|M.ulcerans_Agy99 LQIAGWND--------------------------GGERTR-GVHAYVALT
MMAR_1471|M.marinum_M LQIAGWND--------------------------GGERTR-GVHAYVALT
Mb3181|M.bovis_AF2122/97 LLVAGWNDATDADDLSPASGRYPQRPAPPRLRSSGGERTR-GVHAYVALT
Rv3157|M.tuberculosis_H37Rv LLVAGWNDATDADDLSPASGRYPQRPAPPRLRSSGGERTR-GVHAYVALT
MAV_4045|M.avium_104 LQVAGWND--------------------------GGDRQR-GVHAYVALT
Mflv_4493|M.gilvum_PYR-GCK LIIAGWNDAGD----------------------RLSWGGR-STHIYFALT
Mvan_1873|M.vanbaalenii_PYR-1 LIMAGWNDARD----------------------ELSWGGR-SVHIYFALI
MSMEG_2051|M.smegmatis_MC2_155 LIIAGWNDASR----------------------QTGLAGR-SVQAYLALT
TH_1091|M.thermoresistible__bu LIVAGWNDADQ----------------------RPGLAGR-AVHLYLALT
MAB_2146|M.abscessus_ATCC_1997 LLLAGWNDGAE----------------------APGYGHRPLAQRYMALM
* :***** . * .: *.**
MUL_2471|M.ulcerans_Agy99 LAIESMVLISVVALDVLLFYVFFEAMLIPMYFLIGGFGQ----GRARSGA
MMAR_1471|M.marinum_M LAIESMVLISVVALDVLLFYVFFEAMLIPMYFLIGGFGQ----GRARSGA
Mb3181|M.bovis_AF2122/97 LAIESMVLMSVIALDVLLFYVFFEAMLIPMYFLIGGFGQ----GAGRSRA
Rv3157|M.tuberculosis_H37Rv LAIESMVLMSVIALDVLLFYVFFEAMLIPMYFLIGGFGQ----GAGRSRA
MAV_4045|M.avium_104 LAIESMVLVSVIALDVLLFYVFFEAMLIPMYFLIGGFGA----GSGRSRA
Mflv_4493|M.gilvum_PYR-GCK LAVEGMVLISLVSLDILLFYVFFEAMLIPMYFLIGGFG-----GEHRGRA
Mvan_1873|M.vanbaalenii_PYR-1 LAVEGMVLISLVSLDILLFYVFFEAMLIPMYFLIGGFG-----GADRSRA
MSMEG_2051|M.smegmatis_MC2_155 LAVEGMVLMSLVALDILLFYVFFEAMLIPMYFLIGGFG-----GENRSRA
TH_1091|M.thermoresistible__bu LAVEGMVLMSLIALDVLLFYVFFEAMLIPMYFLIGGFG-----GAHRGRA
MAB_2146|M.abscessus_ATCC_1997 LIVEAMVLLSFSALDVLLFYIFFEAMLIPMYFLIGGFGSTHSDVKQRSRA
* :*.***:*. :**:****:***************** *. *
MUL_2471|M.ulcerans_Agy99 AVKFLLYNLFGGLIMLAAVIGLYVVTAQN-------GSGTFDFRDIVAGV
MMAR_1471|M.marinum_M AVKFLLYNLFGGLIMLAAVIGLYVVTAQN-------GSGTFDFRDIVAGV
Mb3181|M.bovis_AF2122/97 AVKFLLYNLFGGLIMLAAVIGLYVVTAQY-------DSGTFDFREIVAGV
Rv3157|M.tuberculosis_H37Rv AVKFLLYNLFGGLIMLAAVIGLYVVTAQY-------DSGTFDFREIVAGV
MAV_4045|M.avium_104 AVKFLLYNLFGGLIMLAAVIGLYVVTSQH-------GAGTFDFRDIVAGV
Mflv_4493|M.gilvum_PYR-GCK AVKFLLYNLVGGLIMLAAVVGLYVATATSDAFD----AGTFDFRAIVAAV
Mvan_1873|M.vanbaalenii_PYR-1 AVKFLLYNLFGGLIMLASVVGLYVATAGSDAFD----AGTFDFRAIVAAV
MSMEG_2051|M.smegmatis_MC2_155 AVKFLLYNLFGGLIMLAAVIGLYVVTAGSDAFA----AGTFDFREIVAAV
TH_1091|M.thermoresistible__bu AVKFLLYNLFGGLIMLAAVIGLYVVTAGSDALAGPAGGGTFDFRAIVAAV
MAB_2146|M.abscessus_ATCC_1997 AVKFLMYNLFGGLVMLAAVIGLYVVTAQSPAG---EHGGTFDLRDITEMV
*****:***.***:***:*:****.*: .****:* *. *
MUL_2471|M.ulcerans_Agy99 QGGHYGEHPAVFKALFLGFMFAFAIKAPLWPFHRWLPDAAVESTPATAVL
MMAR_1471|M.marinum_M QGGHYGEHPAVFKALFLGFMFAFAIKAPLWPFHRWLPDAAVESTPATAVL
Mb3181|M.bovis_AF2122/97 AAGRYGADPAVFKALFLGFMFAFAIKAPLWPFHRWLPDAAVESTPATAVL
Rv3157|M.tuberculosis_H37Rv AAGRYGADPAVFKALFLGFMFAFAIKAPLWPFHRWLPDAAVESTPATAVL
MAV_4045|M.avium_104 ASGRYGADPAVSKALFLGFMFAFAIKAPLWPFHRWLPDAAVEATPATAVL
Mflv_4493|M.gilvum_PYR-GCK SSGEFAVDPVVMHLLFGGFMFAFAIKAPLWPLHRWLPDSAVEATPASAVL
Mvan_1873|M.vanbaalenii_PYR-1 AGGEFVVNPVVMHLLFGGFMFAFAVKAPLWPFHRWLPDAAVEATPASAVL
MSMEG_2051|M.smegmatis_MC2_155 SSGEFAVNPAIMNFLFLGFMFAFAVKAPLWPFHRWLPDAAVEATPASAVL
TH_1091|M.thermoresistible__bu SAGDLMIDPAVLNLLFLGFMLAFAIKAPLWPFHRWLPDAAVEATPATAVL
MAB_2146|M.abscessus_ATCC_1997 ATGTLGIDPGLAKALFLGFMFAFAVKAPLWPLHTWLPDAAVHATPSSAVL
* .* : : ** ***:***:******:* ****:**.:**::***
MUL_2471|M.ulcerans_Agy99 MMAVMDKVGTFGMLRYCLQLFPDASTYFRPLIVTLAIIGVIYGAVVAIGQ
MMAR_1471|M.marinum_M MMAVMDKVGTFGMLRYCLQLFPDASTYFRPLIVTLAIIGVIYGAVVAIGQ
Mb3181|M.bovis_AF2122/97 MMAVMDKVGTFGMLRYCLQLFPDPSTYFRPLIVTLAIIGVIYGAIVAIGQ
Rv3157|M.tuberculosis_H37Rv MMAVMDKVGTFGMLRYCLQLFPDPSTYFRPLIVTLAIIGVIYGAIVAIGQ
MAV_4045|M.avium_104 MMAVMDKVGTFGMLRYCLQLFPDAATYFRPLIVVLAVIGVIYGAIVAIGQ
Mflv_4493|M.gilvum_PYR-GCK MMAIMDKVGTFGMIRYCLPLFPDSATLFGPAIITLAVIGIVYGAVVAIGQ
Mvan_1873|M.vanbaalenii_PYR-1 MMAIMDKVGTFGMIRYCLPLFPDSASYFSGLIITLAVIGIVYGAVVAIGQ
MSMEG_2051|M.smegmatis_MC2_155 MMAVMDKVGTFGMLRYCLQLFPDASTYFRPVVITLAAIGIVYGAVLAIGQ
TH_1091|M.thermoresistible__bu MMAVMDKVGTFGMLRYSLQLFPDAATLFRPLVVALAVIGIVYGAILAIGQ
MAB_2146|M.abscessus_ATCC_1997 MMAVVDKVGTFAMLRYCVQLFPDASRYFTPVIVTLAVIGIVYGAVVAIGQ
***::******.*:**.: ****.: * ::.** **::***::****
MUL_2471|M.ulcerans_Agy99 TDIMRLIAYTSISHFGFIIAGIFVMTTQGQSGSTLYMLNHGLSTAAVFLI
MMAR_1471|M.marinum_M TDIMRLIAYTSISHFGFIIAGIFVMTTQGQSGSTLYMLNHGLSTAAVFLI
Mb3181|M.bovis_AF2122/97 TDMMRLIAYTSISHFGFIIAGIFVMTTQGQSGSTLYMLNHGLSTAAVFLI
Rv3157|M.tuberculosis_H37Rv TDMMRLIAYTSISHFGFIIAGIFVMTTQGQSGSTLYMLNHGLSTAAVFLI
MAV_4045|M.avium_104 ADVMRLIAYTSISHFGFIILGIFVMTSQGQSGSTLYMLNHGLSTAAVFLI
Mflv_4493|M.gilvum_PYR-GCK TDVMRLIAYTSISHFGFIILGIFVMTSQGQSGSTLYMVNHGISTAALFLI
Mvan_1873|M.vanbaalenii_PYR-1 TDVMRLIAYTSISHFGFIILGIFVMTTQGQSGSTLYMVNHGISTAALFLI
MSMEG_2051|M.smegmatis_MC2_155 TDVMRLIAYTSISHFGFIILGIFVMTSQGQSGSTLYMINHGISTAALFLI
TH_1091|M.thermoresistible__bu TDVMRLIAYTSISHFGFIILGIFVMTSQGQSGSTLYMINHGLSTAALFLI
MAB_2146|M.abscessus_ATCC_1997 TDVMRLIAYTSISHFGFIILGIFAMTSQGQSGSTLYMVNHGISTAALMLV
:*:**************** ***.**:**********:***:****::*:
MUL_2471|M.ulcerans_Agy99 AGFLISRHG-SRAIADYGGVQKVAPVLAGTFLVSAMATLSLPGLAPFISE
MMAR_1471|M.marinum_M AGFLISRHG-SRAIADYGGVQKVAPVLAGTFLVSAMATLSLPGLAPFISE
Mb3181|M.bovis_AF2122/97 AGFLIARRD-SRSIADYGGVQKVAPILAGTFMVSAMATVSLPGLAPFISE
Rv3157|M.tuberculosis_H37Rv AGFLIARRG-SRSIADYGGVQKVAPILAGTFMVSAMATVSLPGLAPFISE
MAV_4045|M.avium_104 AGFLISRHGGSRAIADYGGVQKVAPVLAGTFLVSAMATLSLPGLAPFVSE
Mflv_4493|M.gilvum_PYR-GCK AGFLVSRRG-SRLISAYGGVQKVAPVLAGTFLIAGLATLSLPGLAPFISE
Mvan_1873|M.vanbaalenii_PYR-1 AGFLVSRRG-SRLINAYGGVQKLAPVLAGTFLVAGLATLSLPGLAPFISE
MSMEG_2051|M.smegmatis_MC2_155 AGFLVSRRG-SRLIDSYGGVQKVAPVLAGTFLVAGLATLSLPGLAPFISE
TH_1091|M.thermoresistible__bu AGFLVTRHG-TRLITDYGGVQKVAPVLAGTFLVAGLATLSLPGLAPFISE
MAB_2146|M.abscessus_ATCC_1997 AGFLVTRKG-SRLIASYGGVQKVAPVLAGSFLVAGLATLSLPGLAPFISE
****::*:. :* * ******:**:***:*:::.:**:********:**
MUL_2471|M.ulcerans_Agy99 FLVLLGTFNRYWLAAALGVTALVLSAIYMLWLYQRVMTGPLA----KGNE
MMAR_1471|M.marinum_M FLVLLGTFNRYWLAAAFGVTALVLSAIYMLWLYQRVMTGPLA----KGNE
Mb3181|M.bovis_AF2122/97 FLVLLGTFSRYWLAAAFGVTALVLSAVYMLWLYQRVMTGPIA----EGNE
Rv3157|M.tuberculosis_H37Rv FLVLLGTFSRYWLAAAFGVTALVLSAVYMLWLYQRVMTGPVA----EGNE
MAV_4045|M.avium_104 FLVLLGTFNRYWLAAAFGVTALVLSAVYMLWLYQRMMTGPVA----GGNE
Mflv_4493|M.gilvum_PYR-GCK FLVLIGTFTRYPVFAVLAASALVLSAIYILWMYQRMMTGPVPPSLSEGEN
Mvan_1873|M.vanbaalenii_PYR-1 FLVLIGTFTRYPVFAVLAASALVLSAIYVLWMYQRMMTGPAPLSLVEGEH
MSMEG_2051|M.smegmatis_MC2_155 FLVLIGTFTRYPVVAVFAATALVLSAVYILWTYQRMMTGPVRDGIGDGDR
TH_1091|M.thermoresistible__bu FLVLIGTFTRYPVLAVLAATALVLSAIYILWTYQRMMAGPVR----EDNR
MAB_2146|M.abscessus_ATCC_1997 FLVLVGTFTRYPVAAACAVVALVLAAIYVLWMYQRMMTGPLAP----GNE
****:***.** : *. .. ****:*:*:** ***:*:** .:.
MUL_2471|M.ulcerans_Agy99 RIGDLVPREMIVVAPLIAMLLILGVYPKPVLDVINPAVEHTMSTIGQHDP
MMAR_1471|M.marinum_M RIGDLVPREMIVVAPLIAMLLILGVYPKPVLDVINPAVEHTMSTIGQHDP
Mb3181|M.bovis_AF2122/97 RIGDLVGREMIVVAPLIALLLVLGVYPKPVLDIINPAVENTMTTIGQHDP
Rv3157|M.tuberculosis_H37Rv RIGDLVGREMIVVAPLIALLLVLGVYPKPVLDIINPAVENTMTTIGQHDP
MAV_4045|M.avium_104 KIGDLRARELVVVAPLIALLLVLGIYPKPVLDIINPAVENTMTTIGQHDP
Mflv_4493|M.gilvum_PYR-GCK RIRDLVPRELVVVTPLIALLLLLGIYPKPALDIIDPAVAHTLTTVEQTDP
Mvan_1873|M.vanbaalenii_PYR-1 KIRDLVPREIAVVAPLIALLLVLGVYPKPALDVINPAVTHTLTTIEQPDP
MSMEG_2051|M.smegmatis_MC2_155 PVRDLVPRELVVVAPLLALLLVLGIYPKPALDVINPAVEHTLTTIGQTDP
TH_1091|M.thermoresistible__bu SIRDLIPRELVVVAPLIALLVGLGVYPKPALDIIDPAVGHTLTSIGEHDP
MAB_2146|M.abscessus_ATCC_1997 TIGDLKRRELTVVAPLVALLLVLGIYPKPLLDIVNPAVEQTLRTVNEKDP
: ** **: **:**:*:*: **:**** **:::*** :*: :: : **
MUL_2471|M.ulcerans_Agy99 APKVPHPVSGVSAPRTAEG--PHE-----
MMAR_1471|M.marinum_M APKVPHPVSGVSAPRTAEG--PHE-----
Mb3181|M.bovis_AF2122/97 APSVAHPVPAVGASRTAEG--PHP-----
Rv3157|M.tuberculosis_H37Rv APSVAHPVPAVGASRTAEG--PHP-----
MAV_4045|M.avium_104 APSVP-----MGTPRTAEG--RR------
Mflv_4493|M.gilvum_PYR-GCK ------------APRMAEG--PVR-----
Mvan_1873|M.vanbaalenii_PYR-1 ------------APRMAEG--PVR-----
MSMEG_2051|M.smegmatis_MC2_155 E--------PTVPPTIAEG--AR------
TH_1091|M.thermoresistible__bu ------------RPTITAGDDPVPAGKGR
MAB_2146|M.abscessus_ATCC_1997 PPTVAD-----IALRHGEG--EQR-----
*