For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KIRNILPILVCTTCAVAMTACSSSVNNNADPSDTAAPATNVEVDDGGCTENHDQIAKLQSDLSKNLYGIA AAESGKEERTNPNPIPESDPIKITFSVEGLSHPFLVKQKQLAEETGKRLGAQVNVISANDDVNQQFNDIQ TAIAQGTDALMMMPATTQGLDAVLKQAEAAQIPYFFSQKGMLGVEPASQVLAPYSNEGQQLGEWVVEHYK GQEDVKVAIISGIPGDQSSDARVNSFLLPLLRSCNFNVVANQPGNYRRGDSEKAAQNMLAANPDIDLLFG ANDEAALGGIAALKSSGRQGVDVVGLDGQTDMFAAIQSGQALATVIHKPTAGIVVEEAVDYLRGKPVPEY RVLDEDLVTKETIETGAQPAF*
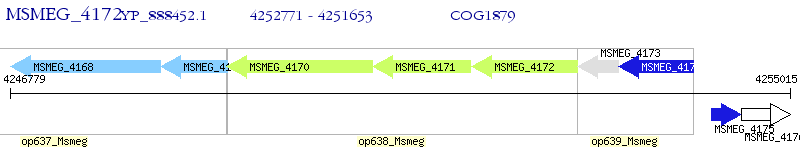
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4172 | - | - | 100% (372) | D-ribose-binding periplasmic protein RbsB |
| M. smegmatis MC2 155 | MSMEG_1712 | - | 8e-23 | 23.99% (296) | ABC transporter periplasmic-binding protein YtfQ |
| M. smegmatis MC2 155 | MSMEG_3095 | - | 7e-22 | 28.73% (275) | D-ribose-binding periplasmic protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00398 | - | 1e-14 | 25.85% (236) | putative D-ribose-binding protein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4172|M.smegmatis_MC2_155 MKIRNILPILVCTTCAVAMTACSSSVNNNADPSDTAAPATNVEVDDGGCT
MLBr_00398|M.leprae_Br4923 ----------MSTGTSVPLRVASIMLILGLLP---------AILPACGFS
:.* :*.: ..* : . * . : * :
MSMEG_4172|M.smegmatis_MC2_155 ENHDQIAKLQSDLSKNLYGIAAAESGKEERTNPNPIPESDPIKITFSVEG
MLBr_00398|M.leprae_Br4923 ESHHKLT------------------------------------IGVSYPT
*.*.::: * .*
MSMEG_4172|M.smegmatis_MC2_155 LSHPFLVKQKQLAEETGKRLGAQVNVISANDDVNQQFNDIQTAIAQGTDA
MLBr_00398|M.leprae_Br4923 ANSPFWNAYTRFIDEGSHQLGVYINAVSAEEDEQKQLSDVETLINQGIDG
. ** .:: :* .::**. :*.:**::* ::*:.*::* * ** *.
MSMEG_4172|M.smegmatis_MC2_155 LMMMPATTQGLDAVLKQAEAAQIPYFFSQKG---MLGVEPASQVLAPYSN
MLBr_00398|M.leprae_Br4923 LIVTPQSTSIAPTLLRIATQANIPVVVVDRYPGYAPGQNKNADYVAFLGP
*:: * :*. ::*: * *:** .. :: * : :: :* .
MSMEG_4172|M.smegmatis_MC2_155 EGQQLGEWVVEHYKGQEDVKVAIISGIPGDQSSDARVNSFLLPLLRSCNF
MLBr_00398|M.leprae_Br4923 NNEKAGSGIAEALMAGGGSKFLALGGMPGNSVAQGRK-AGLESALTAVGH
:.:: *. :.* . . *. :.*:**:. ::.* : * . * : ..
MSMEG_4172|M.smegmatis_MC2_155 NVVANQPGNYRRGDSEKAAQNMLAANP--DIDLLFGANDEAALGGIAALK
MLBr_00398|M.leprae_Br4923 RLVQFQYAGDSEDKGLAAAENMLQAHPAGDANAIWCFNDKLCQGAIKAVY
.:* * .. .... **:*** *:* * : :: **: . *.* *:
MSMEG_4172|M.smegmatis_MC2_155 SSGRQGVDVVG-LDGQTDMFAAIQSGQ-ALATVIHKPTAGIVVEEAVDYL
MLBr_00398|M.leprae_Br4923 NANREKEFIFGGMDLTPQAIAAIENGLYTVSLGAHWLEGGFGLAILYRKI
.:.*: :.* :* .: :***:.* ::: * .*: : :
MSMEG_4172|M.smegmatis_MC2_155 RGKPVPEYRVLDEDLVTKETIETGAQPAF---------------------
MLBr_00398|M.leprae_Br4923 HGQDPAERMVKLDLLKVDKSNVAKFKARYIDNTPHYNWKSTTPFDMTITL
:*: .* * : * ..:: : :. :
MSMEG_4172|M.smegmatis_MC2_155 -
MLBr_00398|M.leprae_Br4923 N