For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDITPDESAAVVAASFDNTPDPRLKEVMQSAVRHLHDFVREVRPTNAEWELAIDFLTRTGQACSETRQEF ILLSDILGVSMLVETLNDAGGRLPEGDGLPVTATTVLGPFHMTTSPTRRLGENIDVVGSGMPCVVKGHVT DVSGNPIPGAKLDVWHANEEGFYDVEQPDKQPAGNGRGIFTTDEDGSYWFRTVTPRHYPIPTDGPAGGLV VQAGRAPYRPAHLHFIAEADGYEAVTTHVFMSKSPYIDSDVVFAVKSSLIRDFVEIEDEHQAADYGVRAP FRLVQFDLALAHAD
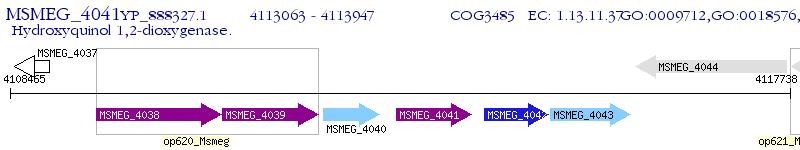
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4041 | - | - | 100% (294) | hydroxyquinol 1,2-dioxygenase |
| M. smegmatis MC2 155 | MSMEG_6713 | - | 2e-90 | 55.09% (285) | hydroxyquinol 1,2-dioxygenase |
| M. smegmatis MC2 155 | MSMEG_1911 | catA | 4e-24 | 36.25% (160) | catechol 1,2-dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1358 | - | 1e-26 | 32.33% (266) | catechol 1,2-dioxygenase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3506 | pcaH | 3e-09 | 30.00% (130) | protocatechuate 3,4-dioxygenase, beta subunit |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4404 | - | 2e-64 | 44.88% (283) | intradiol ring-cleavage dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4041|M.smegmatis_MC2_155 -------MDITPDESAAV------------VAASFDNTPDP---RLKEVM
Mvan_4404|M.vanbaalenii_PYR-1 -------MTMS-DAAIVVEQDRRELDLIDRVQRSFDNCTDP---RLKELM
Mflv_1358|M.gilvum_PYR-GCK MTTIESPINTTSSEASAAASGASATERFRSDKSPFDAVRDTPKERVDLLA
MAV_3506|M.avium_104 ------------------------------------------------MD
:
MSMEG_4041|M.smegmatis_MC2_155 QSAVRHLHDFVREVRPTNAEWELAIDFLTRTGQACSETRQEFILLSDILG
Mvan_4404|M.vanbaalenii_PYR-1 YALVKHLHAFLRETRLTEEEWTRAIEFLTAVGRITDDKRQEFILLSDVLG
Mflv_1358|M.gilvum_PYR-GCK REVLSAVHDTIRRHRVTYDEYNALKAWLISVG-----ADGEWPLFLDVWV
MAV_3506|M.avium_104 PECVASQGDITAEIARIAAQYRADDG-----------GAGQPLLDYPPYR
: . :: : *
MSMEG_4041|M.smegmatis_MC2_155 VSMLVETLNDAGGRLPEGDGLPVTATTVLGPFHMTTSPTRRLGENIDV--
Mvan_4404|M.vanbaalenii_PYR-1 VSMQTIAINNAAYKN-------ATEATVFGPFFAEGSPEVAMGGDISG--
Mflv_1358|M.gilvum_PYR-GCK EHVVEDVATDHREGS---------KGTIEGPYYVPNSPECGARGVIPMR-
MAV_3506|M.avium_104 ATILRHPKQPLVAVDP-------EAAELWAPCFGRDDVDPLDADLTAGHR
: : .* . .
MSMEG_4041|M.smegmatis_MC2_155 -VGSGMPCVVKGHVTDVSGNPIPGAKLDVWHANEEGFY----DVEQPDKQ
Mvan_4404|M.vanbaalenii_PYR-1 -GAAGEPCWVHGSVTDTSGNPVANALIEVWEADAEGFY----DVQYGDGR
Mflv_1358|M.gilvum_PYR-GCK DGEQGTPLVWDGVITSTDGTPLQG-KVELWHADADGFY----SQFAPN-L
MAV_3506|M.avium_104 GEPIGERVVVAGRVVDEAGRPVAGQLVEIWQANAAGRYRHQRDRHPAPLD
* * :.. * *: . :::*.*: * * .
MSMEG_4041|M.smegmatis_MC2_155 PAGNGRGIFTTDEDGSYWFRTVTPRHYPIPTDGPAGGLVVQAGRAPYRPA
Mvan_4404|M.vanbaalenii_PYR-1 TAA--RAHLYTGADGTYCFWAITPTPYPIPHDGPVGEMLEAVGRSPMRAS
Mflv_1358|M.gilvum_PYR-GCK PEWNLRGSFSTGADGVFEINTIRPAPYQIPTDGSCGKLIAAAGWHAWRPA
MAV_3506|M.avium_104 PNFTGAGRCLTGPDGWYRFLTIKPGPYPWRNH-----------HNAWRPA
. . *. ** : : :: * * . . *.:
MSMEG_4041|M.smegmatis_MC2_155 HLHFIAEADGY-EAVTTHVFMSKSPYIDSDVVFA--VKSSLIRDFVEIED
Mvan_4404|M.vanbaalenii_PYR-1 HLHFMVTAPGL-RRLVTHIFVRGDEYLGSDTVFG--VKDSLVKDFQRPST
Mflv_1358|M.gilvum_PYR-GCK HLHVKVSAPGH-ELLTAQLYFPGDPHNDDDIAGA--VKPELILNPTEQAD
MAV_3506|M.avium_104 YIHFSVFGTAFTQRLITQMYFPGDPMFELDPIFQSILDPAARRRLIAHYD
::*. . . . . : ::::. . * :.
MSMEG_4041|M.smegmatis_MC2_155 EHQAADYGVRAPFRLVQFDLALAHAD---------
Mvan_4404|M.vanbaalenii_PYR-1 TSAPGGRSIDGPWSAVQFDIVLAPAEA--------
Mflv_1358|M.gilvum_PYR-GCK GSQR-----------VVYNFVLDPAKA--------
MAV_3506|M.avium_104 HDLTQPEYATG----YRWDIVLAGGGRTPTGAGND
:::.* .