For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTFEAPHIEKIELAEKAGKATAAASGASATERFHLDKSPFDAVRDVPAERVDLLAREVLDAVHATIRRH KVTYEEYNALKAWLINVGEDGEWPLFLDVWVEHVVEEVATAHRHGNKGSIEGPYYVPDSPELGADATMPV RPGEAGTPLVWEGTITSTDGRPLAGKVELWHADSEGFYSQFAPGIPEWNLRGTISTGDDGAFRITTMRPA PYQIPTDGSCGKLIAAAGWHAWRPAHLHVKVSAPGHELLTAQLYFPGDPHNGDDIATAVKPELMLDPQPQ PDGSEKVRYDFVLDPES
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
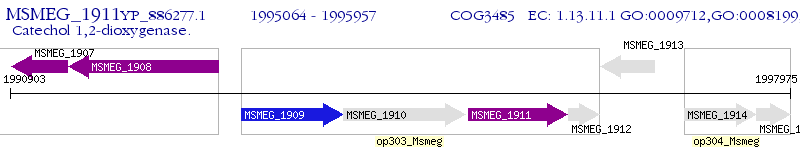
| M. smegmatis MC2 155 | MSMEG_1911 | catA | - | 100% (297) | catechol 1,2-dioxygenase |
| M. smegmatis MC2 155 | MSMEG_6713 | - | 1e-30 | 31.52% (276) | hydroxyquinol 1,2-dioxygenase |
| M. smegmatis MC2 155 | MSMEG_4041 | - | 4e-24 | 36.25% (160) | hydroxyquinol 1,2-dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1358 | - | 1e-137 | 77.63% (295) | catechol 1,2-dioxygenase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3506 | pcaH | 3e-13 | 34.53% (139) | protocatechuate 3,4-dioxygenase, beta subunit |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4404 | - | 3e-26 | 31.34% (268) | intradiol ring-cleavage dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1911|M.smegmatis_MC2_155 MTTFEAPHIEKIELAEKAGKATAAASGASATERFHLDKSPFDAVRDVPAE
Mflv_1358|M.gilvum_PYR-GCK MTTIESP------INTTSSEASAAASGASATERFRSDKSPFDAVRDTPKE
Mvan_4404|M.vanbaalenii_PYR-1 --------------MTMSDAAIVVEQDRRELDLIDRVQRSFDNCTDP---
MAV_3506|M.avium_104 ---------------MDPECVASQGDITAEIARIAAQYRADDGGAGQP--
. . . : . * .
MSMEG_1911|M.smegmatis_MC2_155 RVDLLAREVLDAVHATIRRHKVTYEEYNALKAWLINVG-----EDGEWPL
Mflv_1358|M.gilvum_PYR-GCK RVDLLAREVLSAVHDTIRRHRVTYDEYNALKAWLISVG-----ADGEWPL
Mvan_4404|M.vanbaalenii_PYR-1 RLKELMYALVKHLHAFLRETRLTEEEWTRAIEFLTAVGRITDDKRQEFIL
MAV_3506|M.avium_104 ---LLDYPPYRATILRHPKQPLVAVDPEAAELWAPCFG------------
* . :. : : .*
MSMEG_1911|M.smegmatis_MC2_155 FLDVW--VEHVVEEVATAHRHGNKGSIEGPYYVPDSPELGADATMPVRPG
Mflv_1358|M.gilvum_PYR-GCK FLDVW--VEHVVEDVATDHREGSKGTIEGPYYVPNSPECGARGVIPMRDG
Mvan_4404|M.vanbaalenii_PYR-1 LSDVLGVSMQTIAINNAAYKNATEATVFGPFFAEGSPEVAMGGDISG--G
MAV_3506|M.avium_104 --------RDDVDPLDADLTAGHRG------------------------E
. : : . ..
MSMEG_1911|M.smegmatis_MC2_155 EAGTPLVWEGTITSTDGRPLA-GKVELWHADSEGFYSQFAPGI-----PE
Mflv_1358|M.gilvum_PYR-GCK EQGTPLVWDGVITSTDGTPLQ-GKVELWHADADGFYSQFAPNL-----PE
Mvan_4404|M.vanbaalenii_PYR-1 AAGEPCWVHGSVTDTSGNPVANALIEVWEADAEGFY-DVQYGD-----GR
MAV_3506|M.avium_104 PIGERVVVAGRVVDEAGRPVAGQLVEIWQANAAGRYRHQRDRHPAPLDPN
* * :.. * *: :*:*.*:: * * . .
MSMEG_1911|M.smegmatis_MC2_155 WNLRGTISTGDDGAFRITTMRPAPYQIPTDGSCGKLIAAAGWHAWRPAHL
Mflv_1358|M.gilvum_PYR-GCK WNLRGSFSTGADGVFEINTIRPAPYQIPTDGSCGKLIAAAGWHAWRPAHL
Mvan_4404|M.vanbaalenii_PYR-1 TAARAHLYTGADGTYCFWAITPTPYPIPHDGPVGEMLEAVGRSPMRASHL
MAV_3506|M.avium_104 FTGAGRCLTGPDGWYRFLTIKPGPYPWRNH-----------HNAWRPAYI
. ** ** : : :: * ** . . *.:::
MSMEG_1911|M.smegmatis_MC2_155 HVKVSAPG-HELLTAQLYFPGDPHNGDD------IATAVKPELMLDPQPQ
Mflv_1358|M.gilvum_PYR-GCK HVKVSAPG-HELLTAQLYFPGDPHNDDD------IAGAVKPELILNPTEQ
Mvan_4404|M.vanbaalenii_PYR-1 HFMVTAPG-LRRLVTHIFVRGDEYLGSD------TVFGVKDSLVKDFQRP
MAV_3506|M.avium_104 HFSVFGTAFTQRLITQMYFPGDPMFELDPIFQSILDPAARRRLIAHYDHD
*. * ... . * ::::. ** * ..: *: .
MSMEG_1911|M.smegmatis_MC2_155 PDGSE-----------KVRYDFVLD-----PES----
Mflv_1358|M.gilvum_PYR-GCK ADGSQ-----------RVVYNFVLD-----PAKA---
Mvan_4404|M.vanbaalenii_PYR-1 STTSAPGGRSIDGPWSAVQFDIVLA-----PAEA---
MAV_3506|M.avium_104 LTQPEYAT--------GYRWDIVLAGGGRTPTGAGND
. :::** *