For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SESERSEGFPEGFAGAGSGSLSSEDAAELEALRREAAMLREQLENAVGPQSGLRSARDVHQLEARIDSLA ARNAKLMDTLKEARQQLLALREEVDRLGQPPSGYGVLLATHDDDTVDVFTSGRKMRLTCSPNIEVKELKQ GQTVRLNEALTVVEAGNFEAVGEISTLREILADGHRALVVGHADEERIVWLAEPLVAAKDLPDEPTDYFD DSRPRKLRPGDSLLVDTKAGYAFERIPKAEVEDLVLEEVPDVSYNDIGGLGRQIEQIRDAVELPFLHKDL YKEYSLRPPKGVLLYGPPGCGKTLIAKAVANSLAKKMAEVRGDDAREAKSYFLNIKGPELLNKFVGETER HIRLIFQRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGVEGLENVIVIGASNR EDMIDPAILRPGRLDVKIKIERPDAEAAQDIFSKYLTEDLPVHADDLTEFNGDRALCIKAMIEKVVDRMY AEIDDNRFLEVTYANGDKEVMYFKDFNSGAMIQNVVDRAKKYAIKSVLETGQKGLRIQHLLDSIVDEFAE NEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTESNLGQYL*
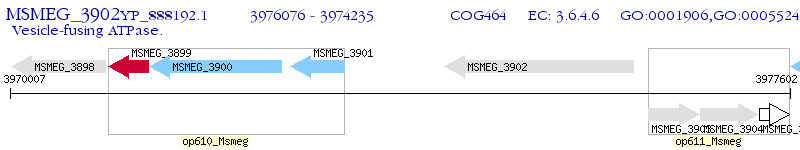
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3902 | - | - | 100% (613) | ATPase, AAA family protein |
| M. smegmatis MC2 155 | MSMEG_0858 | - | 2e-41 | 38.74% (253) | cell division control protein Cdc48 |
| M. smegmatis MC2 155 | MSMEG_6105 | - | 3e-30 | 34.82% (224) | cell division protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2139c | - | 0.0 | 91.22% (615) | ATPase |
| M. gilvum PYR-GCK | Mflv_3069 | - | 0.0 | 89.59% (615) | vesicle-fusing ATPase |
| M. tuberculosis H37Rv | Rv2115c | - | 0.0 | 91.22% (615) | ATPase |
| M. leprae Br4923 | MLBr_01316 | - | 0.0 | 90.23% (614) | putative AAA-family ATPase |
| M. abscessus ATCC 19977 | MAB_2162 | - | 0.0 | 87.34% (616) | ATPase |
| M. marinum M | MMAR_3091 | - | 0.0 | 90.72% (614) | ATPase |
| M. avium 104 | MAV_2400 | - | 0.0 | 90.55% (614) | ATPase, AAA family protein |
| M. thermoresistible (build 8) | TH_2908 | - | 0.0 | 91.55% (592) | PUTATIVE ATPase, AAA family protein |
| M. ulcerans Agy99 | MUL_2337 | - | 0.0 | 90.55% (614) | ATPase |
| M. vanbaalenii PYR-1 | Mvan_3458 | - | 0.0 | 90.75% (616) | vesicle-fusing ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3069|M.gilvum_PYR-GCK -------------------------------------MSESERPEAGDGT
Mvan_3458|M.vanbaalenii_PYR-1 -------------------------------------MSESERSEA---S
MSMEG_3902|M.smegmatis_MC2_155 -------------------------------------MSESERSEG--FP
TH_2908|M.thermoresistible__bu --------------------------------------------------
MMAR_3091|M.marinum_M -------------------------------------MADSERSEA--FG
MUL_2337|M.ulcerans_Agy99 -------------------------------------MADSERSEA--FG
MAV_2400|M.avium_104 MTKILTFRGYLTTHINRQSPVALKYPPRQFGVGKERKMGSSERSEA--FG
MLBr_01316|M.leprae_Br4923 -------------------------------------MGESERSEA--FN
Mb2139c|M.bovis_AF2122/97 -------------------------------------MGESERSEA--FG
Rv2115c|M.tuberculosis_H37Rv -------------------------------------MGESERSEA--FG
MAB_2162|M.abscessus_ATCC_1997 -------------------------------------MSESERSEA----
Mflv_3069|M.gilvum_PYR-GCK DALGASPDTPLSSEDAAELEQLRREASVLREQLADAAGT-IGSARSVRDV
Mvan_3458|M.vanbaalenii_PYR-1 EVFGTSPDSRLSSEDAAELEQLRREAAILREQLEDAGGL-ATSARAARDV
MSMEG_3902|M.smegmatis_MC2_155 EGFAGAGSGSLSSEDAAELEALRREAAMLREQLENAVGP-QSGLRSARDV
TH_2908|M.thermoresistible__bu ----------MSSEDAAELEELRREAAILREQLENAVGA-QGGMRGARDI
MMAR_3091|M.marinum_M T----PDDTPLSSNDAAELEQLRREAAVLREQLESAVGP-QGTARSARDV
MUL_2337|M.ulcerans_Agy99 T----PDDTPLSSNDAAELEQLRREAAVLREQLESAVGP-QGTARSARDV
MAV_2400|M.avium_104 T----PRESDMSSGDEAELEELRREAAMLREQLEHAVGS-HGGARSARDV
MLBr_01316|M.leprae_Br4923 P----PREAGMSSGDIAELEQLRREIVVLREQLEHAVGP-HGSVRSVRDV
Mb2139c|M.bovis_AF2122/97 I----PRDSPLSSGDAAELEQLRREAAVLREQLENAVGS-HAPTRSARDI
Rv2115c|M.tuberculosis_H37Rv I----PRDSPLSSGDAAELEQLRREAAVLREQLENAVGS-HAPTRSARDI
MAB_2162|M.abscessus_ATCC_1997 ----------FGTPDAAEVERLRREVAALREQLEGSAARGPGDSGRLRDL
:.: * **:* **** ***** : . **:
Mflv_3069|M.gilvum_PYR-GCK HQLEARIDSLAARNAKLMDTLKEARQQLLALREEVDRLGQPPSGYGVLLS
Mvan_3458|M.vanbaalenii_PYR-1 HQLEARIDSLAARNAKLMDTLKEARQQLLALREEVDRLGQPPSGYGVLLS
MSMEG_3902|M.smegmatis_MC2_155 HQLEARIDSLAARNAKLMDTLKEARQQLLALREEVDRLGQPPSGYGVLLA
TH_2908|M.thermoresistible__bu HQLEARIDSLAARNAKLMETLKEARQQLLALREEVDRLGQPPSGYGVLLD
MMAR_3091|M.marinum_M HQLEARIDSLAARNSKLMETLKEARQQLLALREEVDRLGQPPSGYGVLLS
MUL_2337|M.ulcerans_Agy99 HQLEARIDSLAARNSKLMETLKEARQQLLALREEVDRLGQPPSGYGVLLS
MAV_2400|M.avium_104 HQLEARIDSLAARNSKLMETLKEARQQLLALREEVDRLGQPPSGYGVLLA
MLBr_01316|M.leprae_Br4923 HQLEARIDSLTARNSKLMDTLKEARQQLLALREEVDRLGQPPSGYGVLLA
Mb2139c|M.bovis_AF2122/97 HQLEARIDSLAARNSKLMETLKEARQQLLALREEVDRLGQPPSGYGVLLA
Rv2115c|M.tuberculosis_H37Rv HQLEARIDSLAARNSKLMETLKEARQQLLALREEVDRLGQPPSGYGVLLA
MAB_2162|M.abscessus_ATCC_1997 NQLEARIDSLNARNAKLMDTLKEARQQLLALREEVDRLGQPPSGYGVLLA
:********* ***:***:******************************
Mflv_3069|M.gilvum_PYR-GCK VQDDETVDVFTSGRRMRLTCSPNIDTKGLKKGQTVRLNEALTVVEAGHFE
Mvan_3458|M.vanbaalenii_PYR-1 VQDDETVDVFTSGRKMRLTCSPNIETKELKKGQTVRLNEALTVVEAGHFE
MSMEG_3902|M.smegmatis_MC2_155 THDDDTVDVFTSGRKMRLTCSPNIEVKELKQGQTVRLNEALTVVEAGNFE
TH_2908|M.thermoresistible__bu IQEDGTVDVFTSGRKMRLTCSPNIDVKTLKKGQTVRLNEALTVVEAGAFE
MMAR_3091|M.marinum_M THDDDTVDVFTSGRKMRLTCSPNIEISLLRKGQTVRLNEALTVVEAGTFE
MUL_2337|M.ulcerans_Agy99 AHDDDTVDVFTSGRKMRLTCSPNIEISLLRKGQTVRLNEALTVVEAGTFE
MAV_2400|M.avium_104 THEDDTVDVFTSGRKMRLTCSPNIDVSLLRKGQTVRLNEALTVVEAGTFE
MLBr_01316|M.leprae_Br4923 AHDDETVDVFTSGRKMRLTCSPNIEVASLRKGQTVRLNEALTVVEAGTFE
Mb2139c|M.bovis_AF2122/97 THDDDTVDVFTSGRKMRLTCSPNIDAASLKKGQTVRLNEALTVVEAGTFE
Rv2115c|M.tuberculosis_H37Rv THDDDTVDVFTSGRKMRLTCSPNIDAASLKKGQTVRLNEALTVVEAGTFE
MAB_2162|M.abscessus_ATCC_1997 VQTDETVDVFTSGRKMRVTCSPNIETSELRRGQTVRLNEALTVVEAGNFE
: * *********:**:******: *::**************** **
Mflv_3069|M.gilvum_PYR-GCK SVGEISTLREILADGHRALVVGHADEERIVWLAEPLVASADLPDG---YD
Mvan_3458|M.vanbaalenii_PYR-1 SVGEISTLREILSDGHRALVVGHADEERIVWLAEPLVAAEHLPEGSYADE
MSMEG_3902|M.smegmatis_MC2_155 AVGEISTLREILADGHRALVVGHADEERIVWLAEPLVAAKDLPDEPTDYF
TH_2908|M.thermoresistible__bu SVGEISTLREVLADGHRALVVGHADEERIVWLAEPLVAPEHVKDD-AELT
MMAR_3091|M.marinum_M SVGEISTLREVLADGHRALVVGHADEERIVWLAEPLVAEDLPDGFPDALN
MUL_2337|M.ulcerans_Agy99 SVGEISTLREVLADGHRALVVGHADEERIVWLAEPLVAEDLPDGFPDALN
MAV_2400|M.avium_104 SVGEISTLRELLADGHRALVVGHADEERIVWLAEPLVADNLPDGHPDALN
MLBr_01316|M.leprae_Br4923 AVGEVSTLREVLADGHRALVVGHADEERIVCLAEPLVAENLLDGVPGALN
Mb2139c|M.bovis_AF2122/97 AVGEISTLREILADGHRALVVGHADEERVVWLADPLIAEDLPDGLPEALN
Rv2115c|M.tuberculosis_H37Rv AVGEISTLREILADGHRALVVGHADEERVVWLADPLIAEDLPDGLPEALN
MAB_2162|M.abscessus_ATCC_1997 SVGEISTLREVLDDGHRALVVGHADEERIVWLADPLIAPDLSEVSEDADG
:***:*****:* ***************:* **:**:*
Mflv_3069|M.gilvum_PYR-GCK DDLGDDRPRKLRPGDSLLVDTKAGYAFERVPKAEVEDLVLEEVPDVSYND
Mvan_3458|M.vanbaalenii_PYR-1 DDLTDDRPRKLRPGDSLLVDTKAGYAFERIPKAEVEDLVLEEVPDVSYND
MSMEG_3902|M.smegmatis_MC2_155 DDS---RPRKLRPGDSLLVDTKAGYAFERIPKAEVEDLVLEEVPDVSYND
TH_2908|M.thermoresistible__bu EEQ---RSRKLRPGDSLLVDTKAGYAFERIPKAEVEDLVLEEVPDVSYED
MMAR_3091|M.marinum_M DDT---KPRKLRPGDSLLVDTKAGYAFERIPKAEVEDLVLEEVPDVSYED
MUL_2337|M.ulcerans_Agy99 DDT---KPRKLRPGDSLLVDTKAGYAFERIPKAEVEDLVLEEVPDVSYED
MAV_2400|M.avium_104 DDT---RPRKLRPGDSLLVDTKAGYAFERIPKAEVEDLVLEEVPDVSYED
MLBr_01316|M.leprae_Br4923 DDS---RPRKLRPGDSLLVDPKAGYAFERVPKAEVEDLVLEEVPDVSYQD
Mb2139c|M.bovis_AF2122/97 DDT---RPRKLRPGDSLLVDTKAGYAFERIPKAEVEDLVLEEVPDVSYAD
Rv2115c|M.tuberculosis_H37Rv DDT---RPRKLRPGDSLLVDTKAGYAFERIPKAEVEDLVLEEVPDVSYAD
MAB_2162|M.abscessus_ATCC_1997 ADL---KPRKLRPGDSLLVDTKAGYAFERIPKAEVEDLVLEEVPDVSYSD
: :.************.********:****************** *
Mflv_3069|M.gilvum_PYR-GCK IGGLGRQIEQIRDAVELPFLHKELYREYSLRPPKGVLLYGPPGCGKTLIA
Mvan_3458|M.vanbaalenii_PYR-1 IGGLGRQIEQIRDAVELPFLHKELYREYSLRPPKGVLLYGPPGCGKTLIA
MSMEG_3902|M.smegmatis_MC2_155 IGGLGRQIEQIRDAVELPFLHKDLYKEYSLRPPKGVLLYGPPGCGKTLIA
TH_2908|M.thermoresistible__bu IGGLSRQIEQIRDAVELPFLHGDLYKEYSLRPPKGVLLYGPPGCGKTLIA
MMAR_3091|M.marinum_M IGGLTRQIEQIRDAVELPFLHKELYREYALRPPKGVLLYGPPGCGKTLIA
MUL_2337|M.ulcerans_Agy99 IGGLTRQIEQIRDAVELPFLHKELYREYALRPPKGVLLYGPPGCGKTLIA
MAV_2400|M.avium_104 IGGLTRQIEQIRDAVELPFLHKELYREYALRPPKGVLLYGPPGCGKTLIA
MLBr_01316|M.leprae_Br4923 IGGLTRQIEQIRDAVELPFLHKELYREYALRPPKGVLLYGPPGCGKTLIA
Mb2139c|M.bovis_AF2122/97 IGGLSRQIEQIRDAVELPFLHKELYREYSLRPPKGVLLYGPPGCGKTLIA
Rv2115c|M.tuberculosis_H37Rv IGGLSRQIEQIRDAVELPFLHKELYREYSLRPPKGVLLYGPPGCGKTLIA
MAB_2162|M.abscessus_ATCC_1997 IGGLARQIEQIRDAVELPFLHKDLYQEYSLRPPKGVLLYGPPGCGKTLIA
**** **************** :**:**:*********************
Mflv_3069|M.gilvum_PYR-GCK KAVANSLAKKMAEVRGDDAREAKSYFLNIKGPELLNKFVGETERHIRLIF
Mvan_3458|M.vanbaalenii_PYR-1 KAVANSLAKKMAEVRGDDAREAKSYFLNIKGPELLNKFVGETERHIRLIF
MSMEG_3902|M.smegmatis_MC2_155 KAVANSLAKKMAEVRGDDAREAKSYFLNIKGPELLNKFVGETERHIRLIF
TH_2908|M.thermoresistible__bu KAVANSLAKKMAEVRGENAKEAKSYFLNIKGPELLNKFVGETERHIRLIF
MMAR_3091|M.marinum_M KAVANSLAKKMAEVRGDDSREAKSYFLNIKGPELLNKFVGETERHIRLIF
MUL_2337|M.ulcerans_Agy99 KAVANSLAKKMAEVRGDDSREAKSYFLNIKGPELLNKFVGETERHIRLIF
MAV_2400|M.avium_104 KAVANSLAKKMAEVRGDDAREAKSYFLNIKGPELLNKFVGETERHIRLIF
MLBr_01316|M.leprae_Br4923 KAVANSLAKKMAEVRGDDAREAKSYFLNIKGPELLNKFVGETERHIRLIF
Mb2139c|M.bovis_AF2122/97 KAVANSLAKKMAEVRGDDAHEAKSYFLNIKGPELLNKFVGETERHIRLIF
Rv2115c|M.tuberculosis_H37Rv KAVANSLAKKMAEVRGDDAHEAKSYFLNIKGPELLNKFVGETERHIRLIF
MAB_2162|M.abscessus_ATCC_1997 KAVANSLAKKMAELRGDDSREAKSYFLNIKGPELLNKFVGETERHIRLIF
*************:**::::******************************
Mflv_3069|M.gilvum_PYR-GCK QRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
Mvan_3458|M.vanbaalenii_PYR-1 QRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
MSMEG_3902|M.smegmatis_MC2_155 QRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
TH_2908|M.thermoresistible__bu QRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
MMAR_3091|M.marinum_M QRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
MUL_2337|M.ulcerans_Agy99 QRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
MAV_2400|M.avium_104 QRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
MLBr_01316|M.leprae_Br4923 QRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
Mb2139c|M.bovis_AF2122/97 QRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
Rv2115c|M.tuberculosis_H37Rv QRAREKASEGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
MAB_2162|M.abscessus_ATCC_1997 QRAREKASDGTPVIVFFDEMDSIFRTRGTGVSSDVETTVVPQLLSEIDGV
********:*****************************************
Mflv_3069|M.gilvum_PYR-GCK EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAESAMDIFSKYL
Mvan_3458|M.vanbaalenii_PYR-1 EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAESAQDIFSKYL
MSMEG_3902|M.smegmatis_MC2_155 EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAEAAQDIFSKYL
TH_2908|M.thermoresistible__bu EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAEAAMDIFSKYL
MMAR_3091|M.marinum_M EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAEAAQDIYSKYL
MUL_2337|M.ulcerans_Agy99 EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAEAAQDIYSKYL
MAV_2400|M.avium_104 EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAEAAQDIFSKYL
MLBr_01316|M.leprae_Br4923 EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAEAAQDIYSKYL
Mb2139c|M.bovis_AF2122/97 EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAEAAQDIYSKYL
Rv2115c|M.tuberculosis_H37Rv EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAEAAQDIYSKYL
MAB_2162|M.abscessus_ATCC_1997 EGLENVIVIGASNREDMIDPAILRPGRLDVKIKIERPDAESAQDIFSKYL
****************************************:* **:****
Mflv_3069|M.gilvum_PYR-GCK TDELPIHEDDLSEFSGDRSLTIKAMIEKVVDRMYAEIDDNRFLEVTYANG
Mvan_3458|M.vanbaalenii_PYR-1 TEELPIHDDDLAEFSGDRSLTIKAMIEKVVDRMYAEIDDNRFLEVTYANG
MSMEG_3902|M.smegmatis_MC2_155 TEDLPVHADDLTEFNGDRALCIKAMIEKVVDRMYAEIDDNRFLEVTYANG
TH_2908|M.thermoresistible__bu TPDLPVHADDLAEFGGDREACIKAMIEKVVDRMYAEIDENRFLEVTYANG
MMAR_3091|M.marinum_M TETLPVHADDLAEFEGERPACIKAMIEKVVDRMYAEIDDNRFLEVTYANG
MUL_2337|M.ulcerans_Agy99 TETLPVHADDLAEFEGERPACIKAMIEKVVDRMYAEIDDNRFLEVTYANG
MAV_2400|M.avium_104 TEELPLHADDLAEFGGDRTACIKAMIEKVVERMYAEIDDNRFLEVTYANG
MLBr_01316|M.leprae_Br4923 TESLPVHADDLTEFDGDRAACIKAMIEKVVDRMYAEIDDNRFLEVTYANG
Mb2139c|M.bovis_AF2122/97 TEFLPVHADDLAEFDGDRSACIKAMIEKVVDRMYAEIDDNRFLEVTYANG
Rv2115c|M.tuberculosis_H37Rv TEFLPVHADDLAEFDGDRSACIKAMIEKVVDRMYAEIDDNRFLEVTYANG
MAB_2162|M.abscessus_ATCC_1997 TVDLPVHSEDLAEFGGDRGLTVKAMIEKVVDRMYAEIDDNRFLEVTYANG
* **:* :**:** *:* :********:*******:***********
Mflv_3069|M.gilvum_PYR-GCK DKEVMYFKDFNSGAMIQNVVDRAKKNAIKAVLETGQRGLRIQHLLDSIVD
Mvan_3458|M.vanbaalenii_PYR-1 DKEVMYFKDFNSGAMIQNVVDRAKKNAIKAVLETGQRGLRIQHLLDSIID
MSMEG_3902|M.smegmatis_MC2_155 DKEVMYFKDFNSGAMIQNVVDRAKKYAIKSVLETGQKGLRIQHLLDSIVD
TH_2908|M.thermoresistible__bu DKEVMYFKDFNSGAMIQNVVDRAKKYAIKSVLETGQPGLRIQHLLDSIVD
MMAR_3091|M.marinum_M DKEVMYFKDFNSGAMIQNVVDRAKKNAIKSVLETGQPGLRIQHLLDSIVD
MUL_2337|M.ulcerans_Agy99 DKEVMYFKDFNSGAMIQNVVDRAKKNAIKSVLETGQPGLRIQHLLDSIVD
MAV_2400|M.avium_104 DKEVMYFKDFNSGAMIQNVVDRAKKNAIKSVLETGQPGLRIQHLLDSIVD
MLBr_01316|M.leprae_Br4923 DKEVMYFKDFNSGAMIQNVVDRAKKNAIKSVLETGQPGLRIQHLLDSIVD
Mb2139c|M.bovis_AF2122/97 DKEVMYFKDFNSGAMIQNVVDRAKKNAIKSVLETGQPGLRIQHLLDSIVD
Rv2115c|M.tuberculosis_H37Rv DKEVMYFKDFNSGAMIQNVVDRAKKNAIKSVLETGQPGLRIQHLLDSIVD
MAB_2162|M.abscessus_ATCC_1997 DKEVMYFKDFNSGAMIQNVVDRAKKNAIKSVLETGQKGLRIQHLLDSIVD
************************* ***:****** ***********:*
Mflv_3069|M.gilvum_PYR-GCK EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTES
Mvan_3458|M.vanbaalenii_PYR-1 EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTES
MSMEG_3902|M.smegmatis_MC2_155 EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTES
TH_2908|M.thermoresistible__bu EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKTSSASRAIDTES
MMAR_3091|M.marinum_M EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTES
MUL_2337|M.ulcerans_Agy99 EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTES
MAV_2400|M.avium_104 EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTES
MLBr_01316|M.leprae_Br4923 EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTES
Mb2139c|M.bovis_AF2122/97 EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTES
Rv2115c|M.tuberculosis_H37Rv EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTES
MAB_2162|M.abscessus_ATCC_1997 EFAENEDLPNTTNPDDWARISGKKGERIVYIRTLVTGKSSSASRAIDTES
**************************************:***********
Mflv_3069|M.gilvum_PYR-GCK NLGQYL
Mvan_3458|M.vanbaalenii_PYR-1 NLGQYL
MSMEG_3902|M.smegmatis_MC2_155 NLGQYL
TH_2908|M.thermoresistible__bu NLGQYL
MMAR_3091|M.marinum_M NLGQYL
MUL_2337|M.ulcerans_Agy99 NLGQYL
MAV_2400|M.avium_104 NLGQYL
MLBr_01316|M.leprae_Br4923 NLGQYL
Mb2139c|M.bovis_AF2122/97 NLGQYL
Rv2115c|M.tuberculosis_H37Rv NLGQYL
MAB_2162|M.abscessus_ATCC_1997 SLGQYL
.*****