For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRSIRIVAAAALAIGLLAGCVSRADQSAPQAAPETVPLSELSGLTLQIGDQKGGTEALLRAAGALEDLPY DVAFSTFTSGPPQIEAATAGKIDFAITGNTPPIFGAASNAKVKVVSAYDGGGFGDQLLVHADSPIQEVTD LRGKSIALAKGSSAHGHTLVQLHRAGLTPDDVKLVFLQPADALSAFKQGRVDVWAVWDPYTAQAEKELPV RSIAQATGVTNGAGFGVASAAALADPKRNTALADLLVRYSKAVKWAHDNRDEWVARYSAEVGLDPEVGAV AQGRSLRLPTDLSDELVASEQEMADLFAESGQIASAPDFSQWVDRRFADQLSPLYIERD*
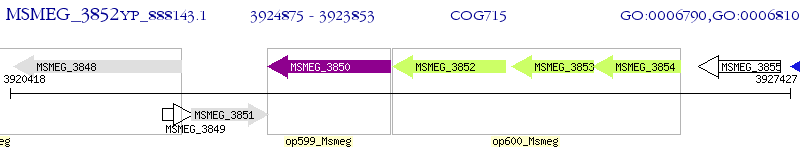
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3852 | - | - | 100% (340) | aliphatic sulfonate binding protein |
| M. smegmatis MC2 155 | MSMEG_0114 | - | 1e-19 | 27.65% (340) | extracellular solute-binding protein, family protein 3 |
| M. smegmatis MC2 155 | MSMEG_0550 | - | 1e-18 | 25.63% (277) | sulfonate binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0723 | - | 1e-13 | 25.96% (312) | substrate-binding region of ABC-type glycine betaine transport |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2218 | - | 1e-109 | 58.48% (330) | sulfonate ABC transporter periplasmic sulfonate-binding protein |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_2435 | - | 1e-129 | 69.18% (331) | sulfonate binding protein |
| M. thermoresistible (build 8) | TH_2996 | - | 2e-28 | 26.41% (337) | PUTATIVE putative aliphatic sulfonates family ABC transporter, |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3411 | - | 1e-134 | 70.71% (338) | aliphatic sulfonate ABC transporter periplasmic ligand-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3852|M.smegmatis_MC2_155 MVRSIRIVA-----------AAALAIG--LLAGCVSRADQSAPQAAPETV
Mvan_3411|M.vanbaalenii_PYR-1 MSAKTTLVARS---------VAVLAVLGLLLTGCVSRDEASGAQEAPETV
MAV_2435|M.avium_104 MIRGV---------------VALTATVGLLLTGCVSRTTTSGPTPPPAAV
MAB_2218|M.abscessus_ATCC_1997 MTPKPRTTEESSLHSWVFKSIVSATAVVLSLTACTTGRDDKGDAAAPALV
TH_2996|M.thermoresistible__bu MHTAIEMLRLT---------LATLISAAALTACGATQSPDSPGLPADAAL
Mflv_0723|M.gilvum_PYR-GCK MRFKAL--------------LVALVAGMLALAGCSVGS--GGQSGNDPEK
* . :
MSMEG_3852|M.smegmatis_MC2_155 PLSELSGLTLQIGDQKGGTEALLRAAGALEDLPYDVAFSTFTSGPPQIEA
Mvan_3411|M.vanbaalenii_PYR-1 PLTELADLTLQIGDQKGGTESLLRAAGVLDDLPYKIAFSTFTSGPPQVEA
MAV_2435|M.avium_104 PLSELSGLTLQVGDQKGGTEALLRAAGQLDNLPYRVAFSTFTSGPPQVEA
MAB_2218|M.abscessus_ATCC_1997 PLSGLSNLTLRVGDQKGGTESLLRAAGELDNVPYKIEFSTFTFGPPQIEA
TH_2996|M.thermoresistible__bu PTEVPPDTTLVVADDANRLRTLLTLSGEQDRLTGEVSYANFSSGPLRLEA
Mflv_0723|M.gilvum_PYR-GCK PTIRIGYQTFPSGDLIVKNNKWLEEALP----DHNIKWVKFDSGADVNTA
* *: .* . * : : : .* *. *
MSMEG_3852|M.smegmatis_MC2_155 ATAGKIDFAITGNTPPIFGAASNAKVKVVSAYDGGGFGDQ--LLVHADSP
Mvan_3411|M.vanbaalenii_PYR-1 ATAGKIDFAITGNTPPIFGAASGAKVKVVAAYDGGGGGDQ--ILVHTDSP
MAV_2435|M.avium_104 ATAGKIDFAITGNTPPIFGAASNARIKAVSAYGGGGAGNR--ILVHADSP
MAB_2218|M.abscessus_ATCC_1997 LTAGKIDFAVTGNTPPIFGAASKARIKVVSGYTNDASGDQ--ILVADASP
TH_2996|M.thermoresistible__bu IRTGNAHLGYVGDVPPILAHYSDADVPIVGAVRRDGTGAL--IATSPDSG
Mflv_0723|M.gilvum_PYR-GCK FVADELDFGALGSSPVARGLSEPLNIPYQVAFILDVAGDNEALVARNAAG
:.: .:. *. * . . : . . * : . :
MSMEG_3852|M.smegmatis_MC2_155 IQEVTDLRGKSIALAKGSSAHGHTLVQLHRAGLTPDDVKLVFLQPADALS
Mvan_3411|M.vanbaalenii_PYR-1 ITEIAELRGKRIAVGKGSSAHGHILAQLKSVGLTPNDVTLVFLQPADALS
MAV_2435|M.avium_104 ITSVSDLRGKAIAVAKGSSSHANLLAQLDRAAIKPADVKFVYLQPADALS
MAB_2218|M.abscessus_ATCC_1997 IRSVADLRGKKVAVGKGSSAHGHLLLQLQKANLTIKDIQPVFLQPADAFT
TH_2996|M.thermoresistible__bu ITSLSDLAGKRIGVNEGTSQQATLLRNLQSVGLGIGDVEPVYLGLAEFAD
Mflv_0723|M.gilvum_PYR-GCK ITTIADLRDKRVATPFASTAHYSLLAALDQNGLSPNDVQLIDLQPQAILA
* :::* .* :. .:: : * *. : *: : *
MSMEG_3852|M.smegmatis_MC2_155 AFKQGRVDVWAVWDPYTAQAEKELPVR---SIAQATGVTNGAGFGVASAA
Mvan_3411|M.vanbaalenii_PYR-1 AFTQREADAWAIWDPYTAQTASQLPVR---SIGQAKDVTNGYWFGVASDQ
MAV_2435|M.avium_104 AFSQHQADAWAIWDPYTAQAEQQIPVR---SIAEAQGVTNGDWIGVASDQ
MAB_2218|M.abscessus_ATCC_1997 ALSQGQADAWAIWDPYTALAQKQLKVR---TLVTATGVSNGYGFGVASQV
TH_2996|M.thermoresistible__bu GLRAEQIQAAVLKQPDRARYLESTAAEGAIEIPNAPGANTGLNYLYAGRD
Mflv_0723|M.gilvum_PYR-GCK AWERGDIDAAYTWLPTLNELRKTGKDLITSRQLAAEGKPTLDLAVVSSEF
. :. * . * . . :.
MSMEG_3852|M.smegmatis_MC2_155 ALADPKRNTALADLLVRYSKAVKWAHDNRDEWVARYSAEVG-LDPEVG-A
Mvan_3411|M.vanbaalenii_PYR-1 ALADPKRNTALADLLVRFEKAARWAQDNPQQWARSYSEAVG-LDLNIA-E
MAV_2435|M.avium_104 ALADPKRNTALGDLLVRFETAVRWARAHPQQWAQSYAAAVG-LDPQVA-A
MAB_2218|M.abscessus_ATCC_1997 ALRDAQRNTALSDVVQRIARASAWARAHPQEWYGKYAAAIG-IDPAAG-E
TH_2996|M.thermoresistible__bu ALIDPARAAAVREFVIAWYRAELWRNGHQDTWISEYLVKDQHVDVDDARK
Mflv_0723|M.gilvum_PYR-GCK AKSHP---EVVDTWRKQQARALDVIADDPAAAAKAIAAEVGLTPEDIEGQ
* .. .: * .
MSMEG_3852|M.smegmatis_MC2_155 VAQGRSLRLPTDLSDELVASE----------QEMADLFAESGQIAS-APD
Mvan_3411|M.vanbaalenii_PYR-1 VSQSRSLRLPTELDDEVVASE----------QKIADLFADAGQIASPAPE
MAV_2435|M.avium_104 VSQARSLRLPTELGDDVVASE----------QKLADLFAAAGQLQS-APR
MAB_2218|M.abscessus_ATCC_1997 LAQSRSLRLPIPLNAEVSASE----------QQLADLFAQSGQIQG-KPT
TH_2996|M.thermoresistible__bu IVESDGVASIPGFTDEVVATQ----------QETIDLLQQAGAFAGRELR
Mflv_0723|M.gilvum_PYR-GCK LTQTVFLTPEQVASAEWLGDDGKPGNLAANLQSASEFLAEQKQIPA-AAP
: : : : . : *. ::: : .
MSMEG_3852|M.smegmatis_MC2_155 FSQWVDRRFADQLSPLYIERD----
Mvan_3411|M.vanbaalenii_PYR-1 FDKWVDRRYNDVLRPLLISTN----
MAV_2435|M.avium_104 FANWVDRRFNAALRPGLVS------
MAB_2218|M.abscessus_ATCC_1997 FSDFIDSRFDNVLKQFFTPSQ----
TH_2996|M.thermoresistible__bu ARDEFDFRFADLTADSPTENRGGQP
Mflv_0723|M.gilvum_PYR-GCK LNTFQDGLYTRGLPGVLTE------
* :