For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TATLVLRFADVGVATYATLRVVGEPARTLKWVLEEPALPAVYSALDDALPDPRGGETVAAAVERSLTAGP FADPKRELKFAKELGSHLIPKPAWNLLIDCVSAPRATLFVTPSPKLARVPWGQLAMPGLHKFRLMELVDV LMAVPPNIVHAPRRSVSWTTRRDNPALLVLDPRIPGQRPDSALGSVLGRPTPGTPLSRHFATAVAQRQVL PRVDGPVDLFRRPDADRGWLARACEHEPSRMLYVGHASAAEGDTADRAALHLAEPEPLSAADVIAQALAV PPRVALLACASGGDYRFDEATGLVAAMVLGGAQLVTATLWPLPTSAGYRTFTADDADPMADTVIGVDLAH NQEQAGIAVNQWQRAQMRRWREGEVTASPLYWASLATFAVDGAR*
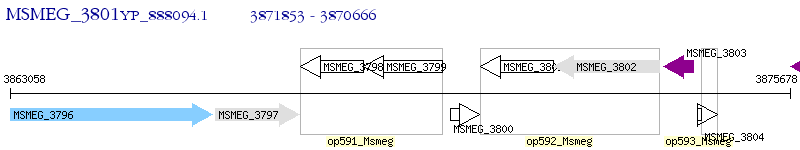
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3801 | - | - | 100% (395) | hypothetical protein MSMEG_3801 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4072c | - | 1e-145 | 63.48% (397) | hypothetical protein MAB_4072c |
| M. marinum M | MMAR_5246 | - | 1e-133 | 54.22% (450) | hypothetical protein MMAR_5246 |
| M. avium 104 | MAV_0375 | - | 1e-135 | 57.40% (453) | hypothetical protein MAV_0375 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3335 | - | 1e-132 | 61.40% (399) | hypothetical protein Mvan_3335 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3801|M.smegmatis_MC2_155 MTA----TLVLRFADVGVATYATLRVVGEPARTLKWVLEEPALPAVYSAL
MAB_4072c|M.abscessus_ATCC_199 MTD----ILVLRFADLGIATYASLRVVGEPSRTVTWVVDQRPLETVCTAL
MMAR_5246|M.marinum_M MNDSDRPTLVLRYADVGITTYASLRVVGQPSRTVNWVVDEPILLAALAEL
MAV_0375|M.avium_104 MTESGRLTLVLRYADLGIATYASLRIVGQPERTVAWVVEEPLLLAALQEL
Mvan_3335|M.vanbaalenii_PYR-1 MTD-AAATLVLRYADVGVATYAGLRVVGQPDSTVTWVIEEPALAEALGLL
*. ****:**:*::*** **:**:* *: **::: * . *
MSMEG_3801|M.smegmatis_MC2_155 DDALPDPRGGETVAAAVERSLTAGPFADPKRELKFAKELGSHLIPKPAWN
MAB_4072c|M.abscessus_ATCC_199 DAALPEATGSETALTAIARALTVGALASPEAELQLARALGAELVAPEGWK
MMAR_5246|M.marinum_M TDALPEPHGNEDRREAIERALCRGPFATRDSELTVAFILGVLLIGTPGWQ
MAV_0375|M.avium_104 AAALPEPHDAENRRAAIGRALSTGAFAAPDTELTLAYILGVLLIGSAGWQ
Mvan_3335|M.vanbaalenii_PYR-1 ADALPDPRPSENVREALERAFTAGSFATATAEQQFADILGRHLISAAGWD
***:. * *: *:: *.:* * .* ** *: .*.
MSMEG_3801|M.smegmatis_MC2_155 LLIDCVSAPRATLFVTPSPKLARVPWGQLAMP--GL--------------
MAB_4072c|M.abscessus_ATCC_199 LLSECVTSPRAVLFVTPSPKLARVPWGQLAMP--GP--------------
MMAR_5246|M.marinum_M LLTECAASPRPVLFISPSARLARVPWGLLALPTAGPTREELVRARQEAIT
MAV_0375|M.avium_104 LLTECVSSPRAVLFVSPSARLARVPWGLLAIPKSGPSKEELVRARQDAIT
Mvan_3335|M.vanbaalenii_PYR-1 LITRHSARQYPALFVAPTARLAQVPWSLLAIP--GD--------------
*: : ..**::*:.:**:***. **:* *
MSMEG_3801|M.smegmatis_MC2_155 --------------------HKFRLMELVDVLMAVPPNIVHAPRRSVSWT
MAB_4072c|M.abscessus_ATCC_199 --------------------DDYRLMELVDVLMSVPPNIVHAPRHPARWR
MMAR_5246|M.marinum_M ASGRAAARIPWQLADITEYTDGYRLMELVDILLAVPQNIVHSARVPARWA
MAV_0375|M.avium_104 AAGRAAAQIPWRLGDLAALTDGHRLMELVEVLMAVPPNIVHAPRTATEWD
Mvan_3335|M.vanbaalenii_PYR-1 --------------------DPRRLLEFADILVAAPPNIANAARTPAQWD
. **:*:.::*::.* **.::.* .. *
MSMEG_3801|M.smegmatis_MC2_155 TRRDNPA-LLVLDPRIPGQRPDSALGSVLGRPTPGTPLSRHFATAVAQRQ
MAB_4072c|M.abscessus_ATCC_199 DRQRGPV-ALLLDPRIPGQRPDSTLGSVLGRPAAEGALTRHFGELMDNHR
MMAR_5246|M.marinum_M ARKDGPP-ILLLDPRVPGQRPDSALGSVLGRPSEETPLARHFAELMRRRP
MAV_0375|M.avium_104 ARRDGPP-LLVLDPRVPGQRPDSALGSVLGRPSPDTPVGRHFAELVRRRP
Mvan_3335|M.vanbaalenii_PYR-1 ERRDNRARLLILDPRVPGQRPDSALGSVLGRPDPDAALTRHFAELRARCE
*: . *:****:*******:******** .: ***. .
MSMEG_3801|M.smegmatis_MC2_155 VLPRVDGPVDLFRRPDADRGWLARACEHEPSRMLYVGHASAAEGDT----
MAB_4072c|M.abscessus_ATCC_199 VLPVAGTPRELFRRSDTDRDWLGGACAQNPARLLYVGHASAAEGAVG---
MMAR_5246|M.marinum_M VLPAVDSPSELFRRRDADRDWLSKLLMQQPSRLLYVGHASSADSE---GG
MAV_0375|M.avium_104 VLPRVDAVVDLFRRRDADRRWLAGLLARSPCRLLYVGHASSADDDHRHGG
Mvan_3335|M.vanbaalenii_PYR-1 VLPDVASTVELFRSAHADRGWLAAQLTREPSRLLFVGHATAADGDVG---
*** . :*** .:** **. :.*.*:*:****::*:.
MSMEG_3801|M.smegmatis_MC2_155 -ADRAALHLAE-------------PEPLSAADVIAQALAVPPRVALLACA
MAB_4072c|M.abscessus_ATCC_199 HAARAALHLAE-------------DHPLTAADLMAAQFPIPPRVALLACS
MMAR_5246|M.marinum_M HADHAALHLACGAAIPGDAPAIGDHRPLTASDLMALRMSMPPRVALLACA
MAV_0375|M.avium_104 RADRAALHLADPAAVPGDANAIGDHRPLTASDLMALRLPMPPRVALLACG
Mvan_3335|M.vanbaalenii_PYR-1 HADRAALHLAD-------------EQPLTAADLMTAQLAIPPRAALLACS
* :****** .**:*:*::: :.:***.*****.
MSMEG_3801|M.smegmatis_MC2_155 SGGDYRFDEATGLVAAMVLGGAQLVTATLWPLPTSAGYRTFTADD-----
MAB_4072c|M.abscessus_ATCC_199 SGGDYRFDEATGLAAAMILGGAELVTATLWSLPTTAAYSQFVSYT-----
MMAR_5246|M.marinum_M SGGDYQFDEATGLVAATILGGAQLVTATLWSLPTTAAYRQFVAEDGDPAD
MAV_0375|M.avium_104 SGGDYQFDEATGLVAAMILNGAQLVTATLWSLPTTAAYRQFAMPARGSPE
Mvan_3335|M.vanbaalenii_PYR-1 SGGDYRFDEATGLVAALILGGALVVTATLWSLPTAAGYRQFCPAADAS--
*****:*******.** :*.** :******.***:*.* *
MSMEG_3801|M.smegmatis_MC2_155 --ADPMADTVIGVDLAHNQ-EQAGIAVNQWQRAQMRRWREGEVTASPLYW
MAB_4072c|M.abscessus_ATCC_199 --ADPMTDTVIGVDHAHRD-EDAGRAVNRWQRAQLRRWRDGDRAASPLHW
MMAR_5246|M.marinum_M PMTDPMTEIVAAIDSAHET-DEAGCAVNRWQRAQMRRWRDGEASASPLYW
MAV_0375|M.avium_104 -SVDPMADLVSAVDTAHDAPADAGCALNRWQRAQMRRWRDGDRVASPLYW
Mvan_3335|M.vanbaalenii_PYR-1 --HDPMAAVVIAVDTAHEA-QHAGRAVNAWQRAQLTRWRAGDDAAAPLYW
***: * .:* ** .** *:* *****: *** *: *:**:*
MSMEG_3801|M.smegmatis_MC2_155 ASLATFAVDGAR
MAB_4072c|M.abscessus_ATCC_199 AAVVSFAVDGAR
MMAR_5246|M.marinum_M GAVVSFAVDGAR
MAV_0375|M.avium_104 AALATFTVDGAR
Mvan_3335|M.vanbaalenii_PYR-1 AALVTFAVDGAR
.::.:*:*****