For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GLTGRPPREIPEPRPRETHGPAKVIAMCNQKGGVGKTTSTINLGASLAEYGRRVLLVDLDPQGALSAGLG VPHYELDNTVHNLLVEPRVSIDDVLIKTRVSGMDLVPSNIDLSAAEIQLVNEVGREQSLARALYPVLDRY DYVLIDCQPSLGLLTINGLACADGVVIPTECEYFSLRGLALLTDTVDKVHDRLNPKLSISGILVTRYDPR TVNAREVMARVVERFGDLVFDTVITRTVRFPETSVAGEPITTWAPKSSGAAAYRSLAREVIHRFGV*
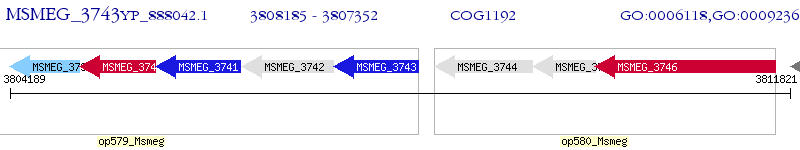
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3743 | soj | - | 100% (277) | SpoOJ regulator protein |
| M. smegmatis MC2 155 | MSMEG_1927 | - | 6e-51 | 44.31% (255) | cobyrinic acid a,c-diamide synthase |
| M. smegmatis MC2 155 | MSMEG_6939 | - | 3e-49 | 40.67% (268) | Soj family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1735 | - | 1e-144 | 88.81% (277) | putative initiation inhibitor protein |
| M. gilvum PYR-GCK | Mflv_3498 | - | 1e-145 | 92.36% (275) | cobyrinic acid a,c-diamide synthase |
| M. tuberculosis H37Rv | Rv1708 | - | 1e-144 | 88.81% (277) | putative initiation inhibitor protein |
| M. leprae Br4923 | MLBr_01367 | - | 1e-142 | 88.77% (276) | putative regulatory protein |
| M. abscessus ATCC 19977 | MAB_2367 | - | 1e-131 | 83.15% (273) | hypothetical protein MAB_2367 |
| M. marinum M | MMAR_2523 | - | 1e-140 | 87.73% (277) | Soj family ATPase |
| M. avium 104 | MAV_3068 | soj | 1e-143 | 89.13% (276) | SpoOJ regulator protein |
| M. thermoresistible (build 8) | TH_3875 | - | 1e-144 | 90.94% (276) | PUTATIVE Cobyrinic acid a,c-diamide synthase |
| M. ulcerans Agy99 | MUL_3237 | - | 1e-140 | 87.00% (277) | Soj family ATPase |
| M. vanbaalenii PYR-1 | Mvan_3279 | - | 1e-146 | 91.67% (276) | cobyrinic acid a,c-diamide synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3498|M.gilvum_PYR-GCK ---------------MTDPTGTPAPVDAPAAPAVAPDPEPTPGLTGRLPR
Mvan_3279|M.vanbaalenii_PYR-1 ---------------MTDPTG--GSLDAPAA---RSDAEPALGLTGRVPR
MSMEG_3743|M.smegmatis_MC2_155 -----------------------------------------MGLTGRPPR
TH_3875|M.thermoresistible__bu ---------------MTD--------DAVTA--------EVVGLTGRPPR
MMAR_2523|M.marinum_M MPAGLPAQADAAVRLSCDVHPDVLHNASRSGMNDHPDSGVSVGLTGRPPR
MUL_3237|M.ulcerans_Agy99 MPAGLPAQADAAVRLSCDVHPDVLHNASRSGMNDHPDSGVSVGLTGRPPR
MAV_3068|M.avium_104 -------------------------------MTERPDNGVELGLTGRPPR
Mb1735|M.bovis_AF2122/97 MPAGLPGQASVAVRLSCDVPPDARHHEPRPGMTDHPDTGNGIGLTGRPPR
Rv1708|M.tuberculosis_H37Rv MPAGLPGQASVAVRLSCDVPPDARHHEPRPGMTDHPDTGNGIGLTGRPPR
MLBr_01367|M.leprae_Br4923 -------------------------------MIDHLDSVVEIGLTGRPPR
MAB_2367|M.abscessus_ATCC_1997 -------------------------MTDGSLDSDFPADDGDLDLTGRPPK
.**** *:
Mflv_3498|M.gilvum_PYR-GCK RIPEPAPRTSHGPAKVIAMCNQKGGVGKTTSTINLGASLAEYGRRVLLVD
Mvan_3279|M.vanbaalenii_PYR-1 QTPEPAPRTSHGPAKVIAMCNQKGGVGKTTSTINLGASLAEYGRRVLLVD
MSMEG_3743|M.smegmatis_MC2_155 EIPEPRPRETHGPAKVIAMCNQKGGVGKTTSTINLGASLAEYGRRVLLVD
TH_3875|M.thermoresistible__bu PIPEPPPKSTHGPAKVIAMCNQKGGVGKTTSTINLGASLAEYGRRVLLVD
MMAR_2523|M.marinum_M AIPDPKPLSSHGPAKVVAMCNQKGGVGKTTSTINLGAALAEYDRRVLLVD
MUL_3237|M.ulcerans_Agy99 AIPDPKPLSSHGPAKVVAMCNQKGGVGKTTSTINLGAALAEYDRRVLLVD
MAV_3068|M.avium_104 AIPEPQPRTSHGPAKVVAMCNQKGGVGKTTSTINLGAALAEYGRRVLLVD
Mb1735|M.bovis_AF2122/97 AIPDPTPRSSHGPAKVIAMCNQKGGVGKTTSTINLGAALGEYGRRVLLVD
Rv1708|M.tuberculosis_H37Rv AIPDPAPRSSHGPAKVIAMCNQKGGVGKTTSTINLGAALGEYGRRVLLVD
MLBr_01367|M.leprae_Br4923 AIPEPRPRSSHGPAKVVAMCNQKGGVGKTTSTINLGAALTEFGRRVLLVD
MAB_2367|M.abscessus_ATCC_1997 DIPEPKPLTSHGPAKVIAMCNQKGGVGKTTSTINLGAALAGYGRRVLLVD
*:* * :******:********************:* :.*******
Mflv_3498|M.gilvum_PYR-GCK LDPQGALSAGLGVPHYELEHTVHNLLVEPRVSIDQVLIKTRVPGLDLVPS
Mvan_3279|M.vanbaalenii_PYR-1 LDPQGALSAGLGVPHYELEHTVHNLLVEPRVSIDQVLIKTRVPGLDLVPS
MSMEG_3743|M.smegmatis_MC2_155 LDPQGALSAGLGVPHYELDNTVHNLLVEPRVSIDDVLIKTRVSGMDLVPS
TH_3875|M.thermoresistible__bu LDPQGALSAGLGVPHYDLDHTVYNLLVEPRVSIDEVLVSTRVRNLDLVPS
MMAR_2523|M.marinum_M MDPQGALSAGLGVAHYELEKTIHNVLVEPRVSVDDVLIHTRVKNLDLVPS
MUL_3237|M.ulcerans_Agy99 MDPQGALSAGLGVAHYELEKTIHNVLVEPRVSVDDVLIHTRVKNLDLVPS
MAV_3068|M.avium_104 MDPQGALSAGLGVPHYELEKTIHNVLVEPRVSIDDVLLQTRVKHMDLVPS
Mb1735|M.bovis_AF2122/97 MDPQGALSAGLGVPHYELDKTIHNVLVEPRVSIDDVLIHSRVKNMDLVPS
Rv1708|M.tuberculosis_H37Rv MDPQGALSAGLGVPHYELDKTIHNVLVEPRVSIDDVLIHSRVKNMDLVPS
MLBr_01367|M.leprae_Br4923 IDPQGALSAGLGVPHYELDRTIHNLMVEPLVSIDDVLIHTRVRYLDLVPS
MAB_2367|M.abscessus_ATCC_1997 LDPQGALSAGLGIAHHELETTVHNLLVEPRVSVDDVLMRTRVEGLDLIPS
:***********:.*::*: *::*::*** **:*:**: :** :**:**
Mflv_3498|M.gilvum_PYR-GCK NIDLSAAEIQLVNEVGREQTLARALYPVLDRYDYVLIDCQPSLGLLTVNG
Mvan_3279|M.vanbaalenii_PYR-1 NIDLSAAEIQLVNEVGREQSLARALYPVLDRYDYVLIDCQPSLGLLTVNG
MSMEG_3743|M.smegmatis_MC2_155 NIDLSAAEIQLVNEVGREQSLARALYPVLDRYDYVLIDCQPSLGLLTING
TH_3875|M.thermoresistible__bu NIDLSAAEIQLVTEVGREQALARALHPVLDRYDYVLIDCQPSLGLLTVNG
MMAR_2523|M.marinum_M NIDLSAAEIQLVNEVGREQTLGRALYPVLDRYDYVLIDCQPSLGLLTVNG
MUL_3237|M.ulcerans_Agy99 NIDLSAAEIQLVNEVGREQTLGRALYPVLDRYDYVLVDCQPSLGLLTVNG
MAV_3068|M.avium_104 NIDLSAAEIQLVNEVGREQTLGRALHPVLDRYDYVLIDCQPSLGLLTVNG
Mb1735|M.bovis_AF2122/97 NIDLSAAEIQLVNEVGREQTLARALYPVLDRYDYVLIDCQPSLGLLTVNG
Rv1708|M.tuberculosis_H37Rv NIDLSAAEIQLVNEVGREQTLARALYPVLDRYDYVLIDCQPSLGLLTVNG
MLBr_01367|M.leprae_Br4923 NIDLSAAEIQLVNEVGREQTLARALHPVLDRYDYVLIDCQPSLGLLTVNG
MAB_2367|M.abscessus_ATCC_1997 NIDLSAAEIQLVNEVGREHSLARALHPVLDRYDYVLIDCQPSLGLLTVNA
************.*****::*.***:**********:**********:*.
Mflv_3498|M.gilvum_PYR-GCK LACSDGVVIPTECEFFSLRGLALLTDTVEKVHDRLNPKLEISGILITRYD
Mvan_3279|M.vanbaalenii_PYR-1 LACSDGVIIPTECEFFSLRGLALLTDTVEKVHDRLNPKLSISGILITRYD
MSMEG_3743|M.smegmatis_MC2_155 LACADGVVIPTECEYFSLRGLALLTDTVDKVHDRLNPKLSISGILVTRYD
TH_3875|M.thermoresistible__bu LACSDGVIIPTECEYFSLRGLALLTDTVDKVRDRLNPKLEISGILVTRYD
MMAR_2523|M.marinum_M LACADGVVIPTECEYFSLRGLALLTDTVEKVRDRLNPRLDISGILLTRYD
MUL_3237|M.ulcerans_Agy99 LACADGVVIPTECEYFSLRGLALLTDTVEKVRDRLNPRLDITGILLTRYD
MAV_3068|M.avium_104 LACSDGVVIPTECEYFSLRGLALLTDTVDKVRDRLNPKLEISGILLTRYD
Mb1735|M.bovis_AF2122/97 LACTDGVIIPTECEFFSLRGLALLTDTVDKVRDRLNPKLDISGILITRYD
Rv1708|M.tuberculosis_H37Rv LACTDGVIIPTECEFFSLRGLALLTDTVDKVRDRLNPKLDISGILITRYD
MLBr_01367|M.leprae_Br4923 LACAEGVVIPTECEFFSLRGLALLTDTVDKVRDRLNPKLEISGILITRYD
MAB_2367|M.abscessus_ATCC_1997 LACSEGVVIPMECAFFSLRGLALLTDTVAKVRDRLNPKLAVSGIVITMFD
***::**:** ** :************* **:*****:* ::**::* :*
Mflv_3498|M.gilvum_PYR-GCK NRTVNAREVMARVVERFGDLVFDTVITRTVRFPETSVAGEPITTWAPKSA
Mvan_3279|M.vanbaalenii_PYR-1 PRTVNSREVMARVVERFGDLVFDTVVTRTVRFPETSVAGEPITTWAPKSA
MSMEG_3743|M.smegmatis_MC2_155 PRTVNAREVMARVVERFGDLVFDTVITRTVRFPETSVAGEPITTWAPKSS
TH_3875|M.thermoresistible__bu PRTVNSREVMARVVERFDDLVFDTVITRTVRFPETSVAGEPITTWAPKST
MMAR_2523|M.marinum_M PRTVNSREVMARVVERFGDLVFDTVITRTVRFPETSVAGEPITTWAPKSA
MUL_3237|M.ulcerans_Agy99 PRTVNSREVMARVVERFGDLVFDTVITRTVRFPETSVAGEPITTWAPKSA
MAV_3068|M.avium_104 PRTVNAREVMARVVERFGDLVFDTVITRTVRFPETSVAGEPITTWAPRST
Mb1735|M.bovis_AF2122/97 PRTVNSREVMARVVERFGDLVFDTVITRTVRFPETSVAGEPITTWAPKSA
Rv1708|M.tuberculosis_H37Rv PRTVNSREVMARVVERFGDLVFDTVITRTVRFPETSVAGEPITTWAPKSA
MLBr_01367|M.leprae_Br4923 PRTVNAREVMARVVERFGDLVFDTVITRTVRFPETSVAGEPITTWAPKSG
MAB_2367|M.abscessus_ATCC_1997 ARTLHAREVMARVIEVFGDQVFHTVITRTVRFPETSVAGEPITTWAPKSS
**:::*******:* *.* **.**:*********************:*
Mflv_3498|M.gilvum_PYR-GCK GAEAYRALAREVIHRFGA
Mvan_3279|M.vanbaalenii_PYR-1 GAEAYRALAREVIHRFGA
MSMEG_3743|M.smegmatis_MC2_155 GAAAYRSLAREVIHRFGV
TH_3875|M.thermoresistible__bu GAEAYRSLAREVIHRFGA
MMAR_2523|M.marinum_M GALAYRVLAREFIDRFGV
MUL_3237|M.ulcerans_Agy99 GALAYRALAREFIDRFGV
MAV_3068|M.avium_104 GAIAYRALAREFIDRFGA
Mb1735|M.bovis_AF2122/97 GALAYRALARELIDRFGM
Rv1708|M.tuberculosis_H37Rv GALAYRALARELIDRFGM
MLBr_01367|M.leprae_Br4923 GARAYRALACEFIDRFGA
MAB_2367|M.abscessus_ATCC_1997 GAQAYISLAREVIDRFET
** ** ** *.*.**