For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGSLVVAVDGPAGTGKSSVSRGLARALGARYLDTGAMYRIATLAVLRAGADLTDPAAIEKAAADAEIGVG SDPDVDAAFLAGEDVSSEIRGDAVTGAVSAVSAVPAVRTRLVDIQRKLATEGGRVVVEGRDIGTVVLPDA DVKIFLTASAEERARRRNAQNVANGLPDDYATVLADVQRRDHLDSTRPVSPLRAADDALVVDTSDMDQAQ VIAHLLDLVTAQAGARR*
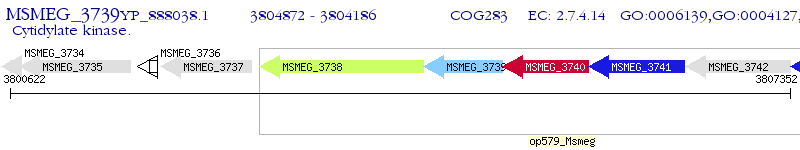
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3739 | cmk | - | 100% (228) | cytidylate kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1739 | cmk | 2e-86 | 73.99% (223) | cytidylate kinase |
| M. gilvum PYR-GCK | Mflv_3494 | cmk | 8e-87 | 72.37% (228) | cytidylate kinase |
| M. tuberculosis H37Rv | Rv1712 | cmk | 2e-86 | 73.99% (223) | cytidylate kinase |
| M. leprae Br4923 | MLBr_01371 | cmk | 7e-78 | 69.82% (222) | cytidylate kinase |
| M. abscessus ATCC 19977 | MAB_2371 | - | 7e-85 | 74.44% (223) | cytidylate kinase (Cmk) |
| M. marinum M | MMAR_2527 | cmk | 3e-84 | 71.88% (224) | cytidylate kinase, Cmk |
| M. avium 104 | MAV_3064 | cmk | 2e-83 | 71.05% (228) | cytidylate kinase |
| M. thermoresistible (build 8) | TH_0201 | cmk | 4e-83 | 73.21% (224) | Probable Cytidylate kinase cmk (CMP kinase) (Cytidine |
| M. ulcerans Agy99 | MUL_3233 | cmk | 1e-83 | 70.98% (224) | cytidylate kinase |
| M. vanbaalenii PYR-1 | Mvan_3275 | cmk | 3e-89 | 74.56% (228) | cytidylate kinase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2527|M.marinum_M MSVGTSGTSVVSAGTSGTSGTNEPAPVGLVVAIDGPAGTGKSSVSRGLAR
MUL_3233|M.ulcerans_Agy99 ----------MSAGTSGTSGTNEPAPVGLVVAIDGPAGTGKSSVSRGLAR
MLBr_01371|M.leprae_Br4923 -------------------------MTDIVVAIDGPAGTGKSSVSRGLAR
MAV_3064|M.avium_104 ------------------------MSADVVVAIDGPAGTGKSSVSKGLAR
Mb1739|M.bovis_AF2122/97 ----------------------MSRLSAAVVAIDGPAGTGKSSVSRRLAR
Rv1712|M.tuberculosis_H37Rv ----------------------MSRLSAAVVAIDGPAGTGKSSVSRRLAR
TH_0201|M.thermoresistible__bu ----------------------------LVVAVDGPAGTGKSSVSKGLAR
MSMEG_3739|M.smegmatis_MC2_155 ------------------------MSGSLVVAVDGPAGTGKSSVSRGLAR
MAB_2371|M.abscessus_ATCC_1997 -----------------------------MVAVDGPSGTGKSSVAKELAR
Mflv_3494|M.gilvum_PYR-GCK ------------------------MSRSLVIAIDGPAGTGKSSVAKGLAR
Mvan_3275|M.vanbaalenii_PYR-1 ------------------------MSRSLVIAIDGPAGTGKSSVARGLAV
::*:***:*******:: **
MMAR_2527|M.marinum_M GLGSRFLDTGAMYRMVTLAVLRAGIDPADEQAVANCAASAQLSVGYDPDA
MUL_3233|M.ulcerans_Agy99 GLGSRFLDTGAMYRMVTLSVLRAGIDPADEQAVADCAASAQLSVGYDPDA
MLBr_01371|M.leprae_Br4923 ELGARYLDTGAMYRMMTLAVLRAGIDPADAAAIGESVWKVQMLSDH----
MAV_3064|M.avium_104 ALGARYLDTGGMYRMVTLAVLRAGIDPADADAVGRAAQAVRMSVDYHPDG
Mb1739|M.bovis_AF2122/97 ELGARFLDTGAMYRIVTLAVLRAGADPSDIAAVETIASTVQMSLGYDPDG
Rv1712|M.tuberculosis_H37Rv ELGARFLDTGAMYRIVTLAVLRAGADPSDIAAVETIASTVQMSLGYDPDG
TH_0201|M.thermoresistible__bu ALGARYLDTGAMYRIVTLAVLRAGVDPGNAEAVAEIADTVQLDVGQSPDD
MSMEG_3739|M.smegmatis_MC2_155 ALGARYLDTGAMYRIATLAVLRAGADLTDPAAIEKAAADAEIGVGSDPDV
MAB_2371|M.abscessus_ATCC_1997 QLGASYLDTGAMYRIVTLWVLRAGVDLTDPAAIAAATDQVPMSVSSDPDA
Mflv_3494|M.gilvum_PYR-GCK SLNASYLDTGAMYRIVTLNALRAGVDLDDAAAIAAATDEVDLAVGYDPDE
Mvan_3275|M.vanbaalenii_PYR-1 AMGARYLDTGAMYRIVTLGVLRAGVDPADAAAVAAVAAEVELAVGHDPGE
:.: :****.***: ** .**** * : *: . . : .
MMAR_2527|M.marinum_M SRFFLSGEDVSAEIRGDDVTRAVSAVSAVSSVRQRLVNLQRAMAEGTGSI
MUL_3233|M.ulcerans_Agy99 SKFFLSGEDVSAEIRGDDVTRAVSAVSAVSSVRQRLLNLQRAMAEGTGSI
MLBr_01371|M.leprae_Br4923 DRYFLGGEDVSSEIRTEEVTQAVSAVSAIPAVRVRLVDLQRQMAEGRGSV
MAV_3064|M.avium_104 DRYFLGGEDVSTEIRGDKVTAAVSAVSSIPAVRTRLVGLQREMASGPGSI
Mb1739|M.bovis_AF2122/97 DSCYLAGEDVSVEIRGDAVTRAVSAVSSVPAVRTRLVELQRTMAEGPGSI
Rv1712|M.tuberculosis_H37Rv DSCYLAGEDVSVEIRGDAVTRAVSAVSSVPAVRTRLVELQRTMAEGPGSI
TH_0201|M.thermoresistible__bu QRFYLGAEDVSDEIRGDAVTRAVSAVSAVPEVRERLVALQRQLAAGEDSV
MSMEG_3739|M.smegmatis_MC2_155 DAAFLAGEDVSSEIRGDAVTGAVSAVSAVPAVRTRLVDIQRKLATEGGRV
MAB_2371|M.abscessus_ATCC_1997 QTALLAGEDVSVPIRGNEVTGAVSAVSAVPAVRERLVRQQRELAESSGAV
Mflv_3494|M.gilvum_PYR-GCK DRFLLAGADVSAEIRGDAVTKAVSAVSAVPAVRARLVEIQRQLAAAAGNV
Mvan_3275|M.vanbaalenii_PYR-1 DRSYLAGEDVSTEIRGDAVTKAVSAVSAVPAVRSRLVALQRELASAPDNV
. *.. *** ** : ** ******::. ** **: ** :* . :
MMAR_2527|M.marinum_M VVEGRDIGTVVFPDARVKIFLTASAETRARRRNDQNVASGLADNYEAVLA
MUL_3233|M.ulcerans_Agy99 VVEGRDIGTVVFPDARVKIFLTASAETRARRRNDQNVASGLADNYEAVLA
MLBr_01371|M.leprae_Br4923 VVEGRDIGTVVLPDAPVKIFLTASPETRARRRNDQNVASGSADDYDRVLA
MAV_3064|M.avium_104 VVEGRDIGTVVLPDAPVKIFLTASAETRARRRNDQNVAAGLADDYEAVLA
Mb1739|M.bovis_AF2122/97 VVEGRDIGTVVFPDAPVKIFLTASAETRARRRNAQNVAAGLADDYDGVLA
Rv1712|M.tuberculosis_H37Rv VVEGRDIGTVVFPDAPVKIFLTASAETRARRRNAQNVAAGLADDYDGVLA
TH_0201|M.thermoresistible__bu VVEGRDIGTVVFPDADVKIFLTATAEERARRRNAQNVAKGLDDDYAGVLA
MSMEG_3739|M.smegmatis_MC2_155 VVEGRDIGTVVLPDADVKIFLTASAEERARRRNAQNVANGLPDDYATVLA
MAB_2371|M.abscessus_ATCC_1997 VVEGRDIGTVVLPDADVKIYLTASAQARAQRRNAQNVSGGGDDEYEKVLA
Mflv_3494|M.gilvum_PYR-GCK VVEGRDIGTVVLPDADVKIFLTASAEVRAQRRTDQNIANGRPGDYESVLA
Mvan_3275|M.vanbaalenii_PYR-1 VVEGRDIGTVVLPAADVKIFLTASAEERARRRDAQNVAAGLPGDYEAVLA
***********:* * ***:***:.: **:** **:: * .:* ***
MMAR_2527|M.marinum_M DVRRRDHLDSTRAVSPLRAAADAIVVDTSEMTEAEVIAHLTELVTQRSGA
MUL_3233|M.ulcerans_Agy99 DVRRRDHLDSTRAVSPLRAAADAIVVDTSEMTEAEVIAHLTELVTQRSGA
MLBr_01371|M.leprae_Br4923 EVRRRDHLDSTRAVSPLYVAQDAMIVDTSKMAEAEVIAHLMDLVKQRSGA
MAV_3064|M.avium_104 EVRRRDHLDSTRRVSPLRAAPDAVVVDTSDMTEAQVIDHLRDLVRQRSEV
Mb1739|M.bovis_AF2122/97 DVRRRDHLDSTRAVSPLQAAGDAVIVDTSDMTEAEVVAHLLELVTRRSEA
Rv1712|M.tuberculosis_H37Rv DVRRRDHLDSTRAVSPLQAAGDAVIVDTSDMTEAEVVAHLLELVTRRSEA
TH_0201|M.thermoresistible__bu DVQRRDRLDSSRAVAPLRAAADAVVVDTSDMTEQQVVEHLTALVAEQAGA
MSMEG_3739|M.smegmatis_MC2_155 DVQRRDHLDSTRPVSPLRAADDALVVDTSDMDQAQVIAHLLDLVTAQAGA
MAB_2371|M.abscessus_ATCC_1997 DVQRRDHLDSTRAVSPLRPAEDALEVDTSDMTQEQVVAHLLDLVRTRAGA
Mflv_3494|M.gilvum_PYR-GCK DVRRRDHLDSTRAVSPLRAAEDAIVVDTSDMNQSEVVARLAELVEQRAGV
Mvan_3275|M.vanbaalenii_PYR-1 DVRRRDHLDSTRAVSPLRAAEDAVVVDTSDMNQAEVVAYLTELVEQKAGV
:*:***:***:* *:** * **: ****.* : :*: * ** :: .
MMAR_2527|M.marinum_M AR
MUL_3233|M.ulcerans_Agy99 AR
MLBr_01371|M.leprae_Br4923 VW
MAV_3064|M.avium_104 AR
Mb1739|M.bovis_AF2122/97 VR
Rv1712|M.tuberculosis_H37Rv VR
TH_0201|M.thermoresistible__bu PR
MSMEG_3739|M.smegmatis_MC2_155 RR
MAB_2371|M.abscessus_ATCC_1997 SR
Mflv_3494|M.gilvum_PYR-GCK SR
Mvan_3275|M.vanbaalenii_PYR-1 AR