For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEIPADLYYTSEHEWVLRTGDDTVRVGITDYAQSALGDVVFVQLPDVGADVASGDAFGEVESTKSVSDLY APVTAKVVAVNGDLEGSPELVNSDPYGEGWLVDLRVEAGTLDEALGGLLDAEGYRAVVTE*
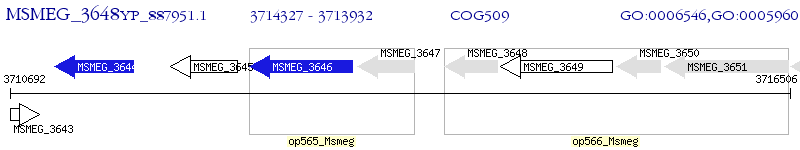
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3648 | gcvH | - | 100% (131) | glycine cleavage system protein H |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1857 | gcvH | 8e-51 | 68.66% (134) | glycine cleavage system protein H |
| M. gilvum PYR-GCK | Mflv_3403 | - | 2e-55 | 77.10% (131) | glycine cleavage system protein H |
| M. tuberculosis H37Rv | Rv1826 | gcvH | 3e-50 | 67.91% (134) | glycine cleavage system protein H |
| M. leprae Br4923 | MLBr_02077 | gcvH | 7e-50 | 68.18% (132) | glycine cleavage system protein H |
| M. abscessus ATCC 19977 | MAB_2402 | - | 1e-51 | 74.60% (126) | glycine cleavage system H protein (GcvH) |
| M. marinum M | MMAR_2703 | gcvH | 4e-51 | 69.40% (134) | glycine cleavage system H protein GcvH |
| M. avium 104 | MAV_2889 | gcvH | 7e-52 | 71.97% (132) | glycine cleavage system protein H |
| M. thermoresistible (build 8) | TH_1125 | gcvH | 1e-54 | 74.81% (131) | PROBABLE GLYCINE CLEAVAGE SYSTEM H PROTEIN GCVH |
| M. ulcerans Agy99 | MUL_3045 | gcvH | 8e-51 | 68.66% (134) | glycine cleavage system protein H |
| M. vanbaalenii PYR-1 | Mvan_3136 | - | 2e-57 | 79.39% (131) | glycine cleavage system protein H |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3403|M.gilvum_PYR-GCK MSEIPADLSYTEEHEWVQRTGDDTVRVGITDYAQAALGDVVFVQLPDVDA
Mvan_3136|M.vanbaalenii_PYR-1 MSEIPADLSYTTEHEWVQRTGDGTVRVGITDYAQSALGDVVFVQLPDVGS
MAB_2402|M.abscessus_ATCC_1997 MTEIPADLHYTEEHEWVRRTGDTTVRIGITDYAQSQLGDVVFVQLPDVGS
MSMEG_3648|M.smegmatis_MC2_155 MSEIPADLYYTSEHEWVLRTGDDTVRVGITDYAQSALGDVVFVQLPDVGA
TH_1125|M.thermoresistible__bu VSDIPSDLYYTSEHEWVRRTGDGTARVGITDYAQNALGDVVFVQLPDVGA
Mb1857|M.bovis_AF2122/97 MSDIPSDLHYTAEHEWIRRSGDDTVRVGITDYAQSALGDVVFVQLPVIGT
Rv1826|M.tuberculosis_H37Rv MSDIPSDLHYTAEHEWIRRSGDDTVRVGITDYAQSALGDVVFVQLPVIGT
MLBr_02077|M.leprae_Br4923 MSDIPSDLHYTAEHEWIRRSREDTVRVGLTDFAQSTLGDVVFVQLPEVGA
MAV_2889|M.avium_104 MSDIPPDLHYTAEHEWVRRSGDDTVRVGITDFAQSALGDVVFVQLPEVGA
MMAR_2703|M.marinum_M MSDIPADLHYTAEHEWIRRTAEDTVRVGITDFAQSALGDVVFVQLPDVGA
MUL_3045|M.ulcerans_Agy99 MSDIPADLHYTAEHEWIRRTAEDTVRVGITDFAQSALGDVVFVQLPDVGA
:::**.** ** ****: *: : *.*:*:**:** ********** :.:
Mflv_3403|M.gilvum_PYR-GCK ALTAGESFGEVESTKSVSDLYAPLTAKVVAVNGDLESNPQLVNSDPYGEG
Mvan_3136|M.vanbaalenii_PYR-1 DVTAGESFGEVESTKSVSDLYAPVTAKVIAVNGDLEGNPQLVNSDPYGEG
MAB_2402|M.abscessus_ATCC_1997 ELTAGSTFGEVESTKSVSDLFAPITAKVIAANGDLDSDPQLVNSDPYGEG
MSMEG_3648|M.smegmatis_MC2_155 DVASGDAFGEVESTKSVSDLYAPVTAKVVAVNGDLEGSPELVNSDPYGEG
TH_1125|M.thermoresistible__bu EITAGESFGEVESTKSVSDLFAPVTAKVVAVNDNLEASPDLVNSDPYGEG
Mb1857|M.bovis_AF2122/97 AVTAGETFGEVESTKSVSDLYAPISGKVSAVNSDLDGTPQLVNSDPYGAG
Rv1826|M.tuberculosis_H37Rv AVTAGETFGEVESTKSVSDLYAPISGKVSEVNSDLDGTPQLVNSDPYGAG
MLBr_02077|M.leprae_Br4923 ELAAGKSFGEVESTKSVSDLYAPVSGTVSAVNTDLEGSPQLVNSDPYGAG
MAV_2889|M.avium_104 QLTAGESFGEVESTKSVSDLYAPVSGTVTAVNTDLEGSPQLVNSDPYGAG
MMAR_2703|M.marinum_M EVTAGETFGEVESTKSVSDLFAPISGKVAAVNADLEATPQLVNTDPYGAG
MUL_3045|M.ulcerans_Agy99 EVTAGETFGEVESTKSVSDLFAPISGKVAAVNADLEATPQLVNTDPYGAG
:::*.:*************:**::..* .* :*:. *:***:**** *
Mflv_3403|M.gilvum_PYR-GCK WLVELQAD--SAS-LDAALAELFDAEGYRGHVAD
Mvan_3136|M.vanbaalenii_PYR-1 WLVELQTE--TES-MQAGLASLLDAEGYRAHVAD
MAB_2402|M.abscessus_ATCC_1997 WLVDLEAA--SAADLDAALNDLLDASGY-GDATD
MSMEG_3648|M.smegmatis_MC2_155 WLVDLRVE--AGT-LDEALGGLLDAEGYRAVVTE
TH_1125|M.thermoresistible__bu WLIDVQAD--PDE-LGAALQNLLTADGYRAVIDE
Mb1857|M.bovis_AF2122/97 WLLDIQVDSSDVAALESALTTLLDAEAYRGTLTE
Rv1826|M.tuberculosis_H37Rv WLLDIQVDSSDVAALESALTTLLDAEAYRGTLTE
MLBr_02077|M.leprae_Br4923 WLLDVHV--SDVGALESAIATLLDAETYRGTLTK
MAV_2889|M.avium_104 WLLDVQV--SDAAALESAITALLDAEAYRGTLTE
MMAR_2703|M.marinum_M WLLDVQVEGSDVAALESALAALLDAQTYRDTVTE
MUL_3045|M.ulcerans_Agy99 WLLDVQVEGSDVAALESALAALLDVQTYRDTVTE
**:::.. : .: *: .. * .