For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQQHEREALSALRLTWAPTADDLWHSQGALHVPGLHERAMTDVLAAFDDAARETDSSPLGVVVRGPAGSG KTHLLGQVRERVQTAGGFFFLVELLDATSFWQSARAGILESLGRPGGERETQLKDLLWELASIAHISRAN RRAIIGDDDLTPEILTDFVTALHKARRQTVKRAHHTLRALVLLGAADLDLQDIGEAFLVGNVGTLGATDG SEAELAAWGLPVPTLTPQESVRDISRLVALAGPAVLALDQIDTLLAQSIERTADRVSGENSEPASLDNRD LEHVAHGLMSVRQTMRRTVSVVACLPAAWEAIENRATASVHDRFRTTALLQGLPTPDIGRAILERRFSAT YSAVGFSAPYPSWPILPSAFDDATQYTPRQLLKRADAHVRHCLEHDTVEELTRLTGEVDTTHGPGHAGGT RTDTSELDRRFAEYRSRAVTAAALDPDGEDTTMPGLLSAALEAWITELGEVEQTFWPDPPPGQRVVLHAR LRQSLDAATDDERHWAFRAVASGNAIAVQNRIRKALEATGLNPERRTLFLLRNTPWPRGAKTAAMVDEFT AAGGRVLPMSDDDIRTMTALRDLIDDNHPDLPGWLRRRRPAHAIDLLRAALGDVAGEAPDLEATDPRMPI ELPSAGSEFVPQPDPAPAVAIEHSPTAITLGVDATGGQPVSVDLAALRKHTAIFAGSGSGKTVLIRRLVE ECALRGVSSIVLDPNNDLSRLGSRWPESPPGWNPADHERSDEYFANTEVVVWTPRRATGRPLSFQPLPDF ASVLDDDDEFSDAVESAVAALEPRALIVGNTAKANRSRAVLREALRYYGASRSVTLSGFIDLLGDLPDDV SALGNARALAAELGQNLRAATVNDPLFGGAGTAADPGVLLTPSAGYRARVSVISMVGLTNEQQREGFVNQ LQMALFAWIKRNPAGNRPLGGLLVMDEAQNFAPSSHTTASTHSTLALSSQARKYGLGLVFATQSPRGLHN HIPGNSATQFYGLLNSPAQIAVAREMARVKGGLVPDISRLRSGQFYLALEGNAFHKIQTPWCLSHHPPSP PTTDEVLRLAQQREPAAP
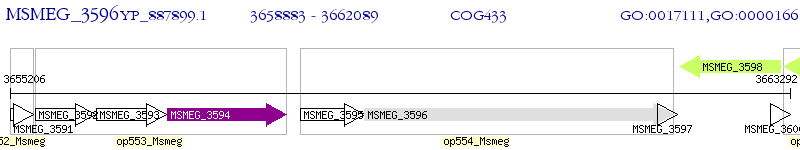
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3596 | - | - | 100% (1068) | ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5149 | - | 0.0 | 64.57% (1064) | AAA ATPase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1183 | - | 0.0 | 65.63% (1062) | AAA ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5149|M.gilvum_PYR-GCK MDTLQREALASLRLTWAPTSDDLWRPQAATHVTGLNEHAVDDVMDAFGDA
Mvan_1183|M.vanbaalenii_PYR-1 MDTVQREALASLRLTWAPTSDDLWRPQGATHVTGLNEHAVDDVMDAFGDA
MSMEG_3596|M.smegmatis_MC2_155 MQQHEREALSALRLTWAPTADDLWHSQGALHVPGLHERAMTDVLAAFDDA
*: :****::********:****:.*.* **.**:*:*: **: **.**
Mflv_5149|M.gilvum_PYR-GCK LRDEASSPMGVVIRGSAGSGKTHLLGQVRERVQAGGGYFFVVELLDAASF
Mvan_1183|M.vanbaalenii_PYR-1 LADPASTPLGVVIRGPAGSGKTHLLGQIRERVQDGGGFFFVVELLDAASF
MSMEG_3596|M.smegmatis_MC2_155 ARETDSSPLGVVVRGPAGSGKTHLLGQVRERVQTAGGFFFLVELLDATSF
: *:*:***:**.***********:***** .**:**:******:**
Mflv_5149|M.gilvum_PYR-GCK WQSVRSGVLESLGRPGTQRETQLKDLLWELSSLAHVSRADRRAIIGDDDL
Mvan_1183|M.vanbaalenii_PYR-1 WESVRSGMLESLGRPGANRETQLKDLLWELASLAHVSRADRRAIIGDDDL
MSMEG_3596|M.smegmatis_MC2_155 WQSARAGILESLGRPGGERETQLKDLLWELASIAHISRANRRAIIGDDDL
*:*.*:*:******** :************:*:**:***:**********
Mflv_5149|M.gilvum_PYR-GCK DAETLYRFIKALSGVAKETVRQCHQVLRALVLLASNDFIAHDIGEGFLQ-
Mvan_1183|M.vanbaalenii_PYR-1 DPETLYRFIKALSSAAKETVRQCHQVLRALVLLGSNDFVAHDIGEGFLE-
MSMEG_3596|M.smegmatis_MC2_155 TPEILTDFVTALHKARRQTVKRAHHTLRALVLLGAADLDLQDIGEAFLVG
.* * *:.** . ::**::.*:.*******.: *: :****.**
Mflv_5149|M.gilvum_PYR-GCK -----GSDEFSIAERGPWGLRSAPESAQMCVRDIVRLVALAGPAVLAVDQ
Mvan_1183|M.vanbaalenii_PYR-1 -----GTEEFTVEERGPWGLRSAPASAQECVRDLSRLVALAGPSVLAVDQ
MSMEG_3596|M.smegmatis_MC2_155 NVGTLGATDGSEAELAAWGLPVPTLTPQESVRDISRLVALAGPAVLALDQ
*: : : * ..*** .. :.* .***: ********:***:**
Mflv_5149|M.gilvum_PYR-GCK IDTLLAQSLAGT----DGDG---------VLEQVAHGLMALRQTTRRTVP
Mvan_1183|M.vanbaalenii_PYR-1 IDTLIAQSVAATGQ-STGDGG-------IVLEQVAHGLMALRQTMRRTVT
MSMEG_3596|M.smegmatis_MC2_155 IDTLLAQSIERTADRVSGENSEPASLDNRDLEHVAHGLMSVRQTMRRTVS
****:***: * *:. **:******::*** ****.
Mflv_5149|M.gilvum_PYR-GCK VVACLPAVWEHIQDRAAASVADRFRVTGMLTPLPSADIGRAILERRFAAS
Mvan_1183|M.vanbaalenii_PYR-1 AVACLPAVWEHIRDQAGAAVADRFRVTGVLTPLPSADIGRAILERRFAAS
MSMEG_3596|M.smegmatis_MC2_155 VVACLPAAWEAIENRATASVHDRFRTTALLQGLPTPDIGRAILERRFSAT
.******.** *.::* *:* ****.*.:* **:.***********:*:
Mflv_5149|M.gilvum_PYR-GCK YRAVGFDPPYPSWPLAPSAFEEAPSYTPRQLLIRADTHVRECLKRGEVVE
Mvan_1183|M.vanbaalenii_PYR-1 YAATGFTPLYPSWPLAPEAFEEAPSYTPRQLLIRADTHVRECLKHGTVVE
MSMEG_3596|M.smegmatis_MC2_155 YSAVGFSAPYPSWPILPSAFDDATQYTPRQLLKRADAHVRHCLEHDTVEE
* *.** . *****: *.**::*..******* ***:***.**::. * *
Mflv_5149|M.gilvum_PYR-GCK LAHLRADAQPDWDTVSTNGSGAVSTALDRRFAEYRRRAVPAAAVAHDGED
Mvan_1183|M.vanbaalenii_PYR-1 LTHLRADAQPTWETQSPNGNAPVSGALDRRFAEYRRRAVPEAALAHDGED
MSMEG_3596|M.smegmatis_MC2_155 LTRLTGEVDTTHGPGHAGGTRTDTSELDRRFAEYRSRAVTAAALDPDGED
*::* .:.:. . ..*. . : ********* ***. **: ****
Mflv_5149|M.gilvum_PYR-GCK VTVPELLSAALTAWIAER-DGEATFVCDPPPGTHVMLHARLRETLDSATD
Mvan_1183|M.vanbaalenii_PYR-1 TTMPELLSAALTAWIAER-DGEQTFVCDPPPGAHVVLHARLRESLDSATD
MSMEG_3596|M.smegmatis_MC2_155 TTMPGLLSAALEAWITELGEVEQTFWPDPPPGQRVVLHARLRQSLDAATD
.*:* ****** ***:* : * ** ***** :*:******::**:***
Mflv_5149|M.gilvum_PYR-GCK DERHWAFRAVAGTNASAVLSRVRKALTASGFGSGGDRRRLFLLRNSPWPS
Mvan_1183|M.vanbaalenii_PYR-1 DERHWAFRAVAATNATAALSRIRKAVTAAGFGAGSDRRSLFLLRNSPWPS
MSMEG_3596|M.smegmatis_MC2_155 DERHWAFRAVASGNAIAVQNRIRKALEATGLNP--ERRTLFLLRNTPWPR
***********. ** *. .*:***: *:*:.. :** ******:***
Mflv_5149|M.gilvum_PYR-GCK GKKTTEEIELFHSAGGRTLPLSDDDIRTMTALRDLFADDDPELAAWLRAR
Mvan_1183|M.vanbaalenii_PYR-1 GPKTAAVIDEFLAAGGRVLPLSDDDVRTMTALRDLIRDDDPDLPGWLRAR
MSMEG_3596|M.smegmatis_MC2_155 GAKTAAMVDEFTAAGGRVLPMSDDDIRTMTALRDLIDDNHPDLPGWLRRR
* **: :: * :****.**:****:*********: *:.*:*..*** *
Mflv_5149|M.gilvum_PYR-GCK RPAHGLTLLREALGGDS------ESAEPAAPEPEP--------DPEDEVV
Mvan_1183|M.vanbaalenii_PYR-1 RPAHGLTLLAEALGDGP------AEPVAVVPEDGPEA-ALPVDEPESVVD
MSMEG_3596|M.smegmatis_MC2_155 RPAHAIDLLRAALGDVAGEAPDLEATDPRMPIELPSAGSEFVPQPDPAPA
****.: ** ***. . . . * * :*:
Mflv_5149|M.gilvum_PYR-GCK ADYDVPADEIPLGLDLQDEKPVTVQLSALRKHVAIFAGSGSGKTVLIRRL
Mvan_1183|M.vanbaalenii_PYR-1 HDYEVPDTELPLGLDVHTEEPVTVGLSALRKHVAIFAGSGSGKTVLIRRL
MSMEG_3596|M.smegmatis_MC2_155 VAIEHSPTAITLGVDATGGQPVSVDLAALRKHTAIFAGSGSGKTVLIRRL
: . :.**:* :**:* *:*****.*****************
Mflv_5149|M.gilvum_PYR-GCK VEECALRGVSSIVLDPNNDLSRLGTAWPQTPAGWRRGDDARADDYLTNTE
Mvan_1183|M.vanbaalenii_PYR-1 VEECALRGVSSIVLDPNNDLSRLGTGWPQPPDGWRRSDGARADDYLRHTE
MSMEG_3596|M.smegmatis_MC2_155 VEECALRGVSSIVLDPNNDLSRLGSRWPESPPGWNPADHERSDEYFANTE
************************: **:.* **. .* *:*:*: :**
Mflv_5149|M.gilvum_PYR-GCK VTVWTPRRTNGRPLAFAPLPDFAGVLDDPDEFTEAVEAACSALEPQALIT
Mvan_1183|M.vanbaalenii_PYR-1 VTIWTPRRTNGRPLAFQPLPDFASVVADYDEFTDAVESACAALEPQALIT
MSMEG_3596|M.smegmatis_MC2_155 VVVWTPRRATGRPLSFQPLPDFASVLDDDDEFSDAVESAVAALEPRALIV
*.:*****:.****:* ******.*: * ***::***:* :****:***.
Mflv_5149|M.gilvum_PYR-GCK GKTQKATRSRAVLREALQHYGKTESPTLGGFIRLLADLPEDVSDIAGAHA
Mvan_1183|M.vanbaalenii_PYR-1 GKTQKATRSRAVLREALQHYGRTPSPTLDGFVRLLADLPEDVSNMAGAHA
MSMEG_3596|M.smegmatis_MC2_155 GNTAKANRSRAVLREALRYYGASRSVTLSGFIDLLGDLPDDVSALGNARA
*:* **.**********::** : * **.**: **.***:*** :..*:*
Mflv_5149|M.gilvum_PYR-GCK IAGDLSQNLRAAMVNDPMFGGAGTPVDPGVLLTPSDGYRARVSVISMIGL
Mvan_1183|M.vanbaalenii_PYR-1 IAGDLSQNLRAAMVNDPLFGGAGTPVDPGVLLTPSDGYRARVSVISMIGL
MSMEG_3596|M.smegmatis_MC2_155 LAAELGQNLRAATVNDPLFGGAGTAADPGVLLTPSAGYRARVSVISMVGL
:*.:*.****** ****:******..********* ***********:**
Mflv_5149|M.gilvum_PYR-GCK PSDDQRQSFVNQLQMALFAWIKRNPAGDRPLGGLLVMDEAQNFAPARGFT
Mvan_1183|M.vanbaalenii_PYR-1 PSDEQRQSFVNQLQMALFAWIKRNPAGERPLGGLLVMDEAQNFAPARGFT
MSMEG_3596|M.smegmatis_MC2_155 TNEQQREGFVNQLQMALFAWIKRNPAGNRPLGGLLVMDEAQNFAPSSHTT
..::**:.*******************:*****************: *
Mflv_5149|M.gilvum_PYR-GCK ACTRSTLALSSQARKYGLGLVFATQAPKGLNLNIPGNAATQFYGLLNAPA
Mvan_1183|M.vanbaalenii_PYR-1 ACTRSTLALSSQARKYGLGLVFATQAPKSLNNNIPGNAATQFYGLLNAPA
MSMEG_3596|M.smegmatis_MC2_155 ASTHSTLALSSQARKYGLGLVFATQSPRGLHNHIPGNSATQFYGLLNSPA
*.*:*********************:*:.*: :****:*********:**
Mflv_5149|M.gilvum_PYR-GCK QIETAREMARAKGGLVDDISRLRAGNFYVATEGESFHRIVAPWCLSHHPA
Mvan_1183|M.vanbaalenii_PYR-1 QIEAAREMARAKGGLVDDISRLRAGNFYVAAEGEAFHRIVAPWCLSYHPS
MSMEG_3596|M.smegmatis_MC2_155 QIAVAREMARVKGGLVPDISRLRSGQFYLALEGNAFHKIQTPWCLSHHPP
** .******.***** ******:*:**:* **::**:* :*****:**.
Mflv_5149|M.gilvum_PYR-GCK SPPSAEEVLNLATSV-----
Mvan_1183|M.vanbaalenii_PYR-1 SPPSAEDVLELAISV-----
MSMEG_3596|M.smegmatis_MC2_155 SPPTTDEVLRLAQQREPAAP
***::::**.** .