For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIELPDAVRAMAARGPRWQTWVDGLAKLVRTQIGDWELTVDGAAMHGYCSIVVPVRDRDGASAVLKLSFP DDETEHEHLALRRWGGHGAVRLLRADPHHRALLLERLSNRNLNELWDIQACEIVADLYRALHVPALPQVR SLPRAVTRWTDGLAALPRSAPVPRRLVEQAITLGTDLATDPSSSGVLIHGDLHYENVLAGPVGSDDGDDT DGGPGSWVAIDPKPFDGDPHYELAPMLWNRWDELTGYVREGVRRRFYAVVDAAGLDADRARAWVIVRMMH NAMWELTERSEPDAHWLTTCVAIAKAVQD
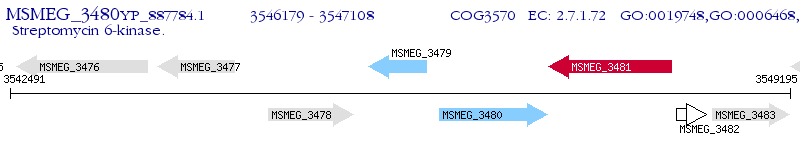
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3480 | - | - | 100% (309) | streptomycin 6-kinase (streptidine kinase) (Streptomycin6-phosphotransferase) (APH(6)) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4430 | - | 1e-76 | 54.74% (274) | aminoglycoside/hydroxyurea antibiotic resistance kinase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3480|M.smegmatis_MC2_155 MIELPDAVRAMAARGPRWQTWVDGLAKLVRTQIGDWELTVDGAAMHGYCS
Mflv_4430|M.gilvum_PYR-GCK ---------MMAA----------------------WSLQADGQEYARTRA
*** *.* .** :
MSMEG_3480|M.smegmatis_MC2_155 IVVPVRDRDGASAVLKLSFPDDETEHEHLALRRWGGHGAVRLLRADPHHR
Mflv_4430|M.gilvum_PYR-GCK YVAPVRTSDGQAATLKIAAPGPGTAHEHLVLRRWDGHGAVRLLRADPHRR
*.*** ** :*.**:: *. * ****.****.*************:*
MSMEG_3480|M.smegmatis_MC2_155 ALLLERLSNRNLNELWDIQACEIVADLYRALHVPALPQVRSLPRAVTRWT
Mflv_4430|M.gilvum_PYR-GCK AILVERAGRDDLTAAPDRRACEIVAQLYRRLHVPAMPQLSSLTEDVRRRT
*:*:** .. :*. * :******:*** *****:**: **.. * * *
MSMEG_3480|M.smegmatis_MC2_155 DGLAALPRSAPVPRRLVEQAITLGTDLATDPSSSGVLIHGDLHYENVLAG
Mflv_4430|M.gilvum_PYR-GCK LALADLPRSAPLPRRLVEQAITLGRELTVEPAER-VVLHGNLHYRNALA-
.** ******:************ :*:.:*:. *::**:***.*.**
MSMEG_3480|M.smegmatis_MC2_155 PVGSDDGDDTDGGPGSWVAIDPKPFDGDPHYELAPMLWNRWDELTGYVRE
Mflv_4430|M.gilvum_PYR-GCK ---------ADREP--WLAISPRPCNGDPPYELAPMLWHRFDDLAGNVRD
:* * *:**.*:* :*** ********:*:*:*:* **:
MSMEG_3480|M.smegmatis_MC2_155 GVRRRFYAVVDAAGLDADRARAWVIVRMMHNAMWELTERSEPDAHWLTTC
Mflv_4430|M.gilvum_PYR-GCK GVRQRFHRLVDGAGFDEDRARAWTLVRVVHAAADDLRTEPASATTQLTRY
***:**: :**.**:* ******.:**::* * :* .. . : **
MSMEG_3480|M.smegmatis_MC2_155 VAIAKAVQD
Mflv_4430|M.gilvum_PYR-GCK VAIAKAIQD
******:**