For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVIDLNADLGESFGVYTYGADAEMMPLITSANIACGGHGGDPAVMRTSVALAHAHDVAVGAHVGLPDRL GFGRREIPTTAQEAYDLCLYQVGALDGFLRAQGTAMQHLKLHGSLYMMANRDRDLAEAVCAAVTALDPAL LIYVLPGSALHTTAVETGLTPVIEIFADRPYRDGVVQMYDRTAELIGGPDQVVTRTLSQLAAISETTGTD VHLTVCVHSDTPGAPALLAAVRSALTAEGHTFRSPTTPASATTRKPILTAALGGE
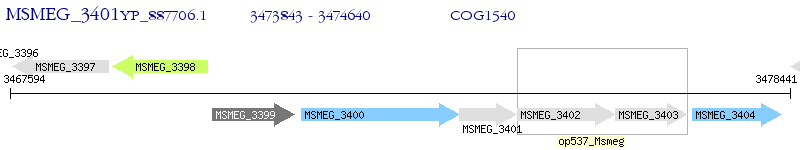
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3401 | - | - | 100% (265) | LamB/YcsF family protein |
| M. smegmatis MC2 155 | MSMEG_6054 | - | 2e-44 | 40.54% (259) | hypothetical protein MSMEG_6054 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1451 | - | 1e-45 | 41.20% (250) | hypothetical protein Mflv_1451 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00333 | - | 6e-41 | 38.87% (247) | LamB/YcsF family protein |
| M. abscessus ATCC 19977 | MAB_0574c | - | 5e-40 | 39.57% (235) | hypothetical protein MAB_0574c |
| M. marinum M | MMAR_5076 | lamB | 1e-41 | 38.40% (250) | lactam utilization protein LamB |
| M. avium 104 | MAV_0580 | - | 8e-40 | 39.60% (250) | hypothetical protein MAV_0580 |
| M. thermoresistible (build 8) | TH_1751 | - | 7e-40 | 39.59% (245) | LamB/YcsF |
| M. ulcerans Agy99 | MUL_4152 | lamB | 7e-42 | 38.40% (250) | LamB/YcsF family protein |
| M. vanbaalenii PYR-1 | Mvan_5327 | - | 5e-44 | 41.63% (245) | hypothetical protein Mvan_5327 |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00333|M.leprae_Br4923 MA--CIDLNADLGEGFGVWRLGDDEAMLRIVTSANVACGFHAGDPAGLLR
MAV_0580|M.avium_104 MA--GIDLNADLGEGFGVWRLGDDDAMLGIVSSANVACGFHAGDPAGLLR
MMAR_5076|M.marinum_M MP--YIDLNADLGEGFGIWQLGDDDAMLGIVTSANVACGFHAGEPAGLLR
MUL_4152|M.ulcerans_Agy99 MP--YIDLNADLGEGFGIWQLGDDDAMLGIVTSANVACGFHAGEPAGLLR
Mflv_1451|M.gilvum_PYR-GCK MS--SVDLNADLGEGFGVWALGDDDAMLDIVTSANVACGFHAGDPATLRR
Mvan_5327|M.vanbaalenii_PYR-1 MTTVSVDLNADLGEGFGVWTLGDDDAMLDIVTSANVACGFHAGDPATLVR
TH_1751|M.thermoresistible__bu VP--TVDLNADLGESFGVWRLGDDEAMLDVITSANVACGFHAGDPAGLIT
MAB_0574c|M.abscessus_ATCC_199 MA--SIDLNADLGEGFGAWRLGDDDAMLDVVTSANLACGFHAGDPATLHR
MSMEG_3401|M.smegmatis_MC2_155 MT--VIDLNADLGESFGVYTYGADAEMMPLITSANIACGGHGGDPAVMRT
:. :********.** : * * *: :::***:*** *.*:** :
MLBr_00333|M.leprae_Br4923 VCRLAAERGVRIGAQVSYRDLVGFGRRFIDVTADDLLADVVYQIGALQAI
MAV_0580|M.avium_104 VCRSAAERGVRVGAQVSYRDLAGFGRRFIDVAADELLADVVYQIGALQAI
MMAR_5076|M.marinum_M VCRAAAERGVRIGAQVGYRDLAGFGRRFIDVDPEDLVAEVVYQIGALQAI
MUL_4152|M.ulcerans_Agy99 VCRAAAERGVRIGAQVGYRDLAGFGRRFIDVDPEDLVAEVVYQIGALQAI
Mflv_1451|M.gilvum_PYR-GCK VCEAAAARGVRIGAQVSYRDLAGFGRRFIDVSSEDLIADVMYQIGALSAL
Mvan_5327|M.vanbaalenii_PYR-1 VCRAAAARGVRIGAQVSYRDLAGFGRRFIDVSAEDLVADVMYQIGALSTL
TH_1751|M.thermoresistible__bu TCRAAAERGVRIGAQVSYRDLAGFGRRFVDADPRDLTADVIYQIGALQAI
MAB_0574c|M.abscessus_ATCC_199 TCHAAATRDIRIGAQVGYRDLAGFGRRFIDIAAADLYADVVYQIGALDAL
MSMEG_3401|M.smegmatis_MC2_155 SVALAHAHDVAVGAHVGLPDRLGFGRREIPTTAQEAYDLCLYQVGALDGF
* :.: :**:*. * ***** : . : :**:***. :
MLBr_00333|M.leprae_Br4923 AQTAGSAVSYVKPHGALYNTIVTNREQGAAVAAAIQLVDSTLPVLGLAGS
MAV_0580|M.avium_104 AHAAGSSVCYVKPHGALYNTIVTHPAQAAAVAEAVRLVDPGLPVLGMAGS
MMAR_5076|M.marinum_M AQSCGSTVSYVKPHGALYNTIVTHRDQAAAVAEAVQMVDATLPVLGMTGS
MUL_4152|M.ulcerans_Agy99 AQSCGSTVSYVKPHGALYNTIVTHRDQAAAVAEAVRMVDATLPVLGMTGS
Mflv_1451|M.gilvum_PYR-GCK AAAAGSSVSYVKPHGALYNAVVTNRLQAHALAAAVHAVDPALPVLGLAGS
Mvan_5327|M.vanbaalenii_PYR-1 AAVAGSSVSYVKPHGALYNAITTNHLQAHAVAEAVHTVDPTLPVLGLAGS
TH_1751|M.thermoresistible__bu AKAAGTRVSYVKPHGALYHSLLSNHAQARAVAEAVHTVDPELPVLSLYGS
MAB_0574c|M.abscessus_ATCC_199 ARTAGSRVSYVKPHGALYNTIIHHEAQADAVARAVRDFNADLPVLTLPHG
MSMEG_3401|M.smegmatis_MC2_155 LRAQGTAMQHLKLHGSLYMMANRDRDLAEAVCAAVTALDPALLIYVLPGS
*: : ::* **:** . . *:. *: .:. * : : .
MLBr_00333|M.leprae_Br4923 TFFDEAARIGLRTVAEAFADRTYRPDGQLISRREPGAVLHDPAVIAQRVV
MAV_0580|M.avium_104 VFFDEAARRGLRVVAEAFADRAYRPDGRLVSRREPGAVLADPAAIAERVL
MMAR_5076|M.marinum_M VFFQQATDLGLRTVAEAFADRAYRSDGQLVSRREHGAVLADAAAIAQRAV
MUL_4152|M.ulcerans_Agy99 VFFQQATDLGLRTVAEAFADRAYRSDGQLVSRREHGAVLADAAAIAQRAV
Mflv_1451|M.gilvum_PYR-GCK VFFGAAEELGLRTVPEAFADRAYRPDGRLVSRRERNSVLHDVDEIAERVI
Mvan_5327|M.vanbaalenii_PYR-1 VFFTAATELGLRTVPEAFADRAYRPDGQLVSRRERNAVLRDVDEIVDRVI
TH_1751|M.thermoresistible__bu VFFAEAHRLGLRTVPEAFADRAYRPDGKLLDRKLRGALLHDEAEIAERVA
MAB_0574c|M.abscessus_ATCC_199 VFGDRARTLGLRVVTEAFADRAYLDDGTLVPRSESGAVLHDPDAVAARIP
MSMEG_3401|M.smegmatis_MC2_155 ALHTTAVETGLTPVIEIFADRPYR-DGVVQMYDRTAELIGGPDQVVTRTL
.: * ** * * ****.* ** : :: . :. *
MLBr_00333|M.leprae_Br4923 TMVTTGKATAVDGTQLAVTVESICLHGDSPNAIQMATAVRDQLNAAGIDI
MAV_0580|M.avium_104 AMVETGAVTAVDGTRLALSVESVCVHGDSPGAVQIATAVRDRLRAAGVEI
MMAR_5076|M.marinum_M SMVASGKVTAVDGTQVPITMESICVHGDSPGAVQIATAVRDRLTAAGNEI
MUL_4152|M.ulcerans_Agy99 SMVASGKVTAVDGTQVPITMESICVHGDSPGAMQIATAVRDRLTAAGNEI
Mflv_1451|M.gilvum_PYR-GCK SMVSRGRVHAVDGSTIPITVESVCVHGDSPGAVQIATAVRKRLVAEGVTL
Mvan_5327|M.vanbaalenii_PYR-1 SMVTTGRVAAVDGSTIPITVESVCVHGDSPGAVQIATAVRERLIADGVTL
TH_1751|M.thermoresistible__bu MMVTTGRVVAVDGSTIPITVESICVHGDTAGAVAIAYAVRDRLRREGVEL
MAB_0574c|M.abscessus_ATCC_199 ALSR--------------TAESICVHGDSPGAVTIARTVRAALAASEIEV
MSMEG_3401|M.smegmatis_MC2_155 SQLA-----AISETTGTDVHLTVCVHSDTPGAPALLAAVRSALTAEGHTF
::*:*.*:..* : :** * .
MLBr_00333|M.leprae_Br4923 RAFC-------------------
MAV_0580|M.avium_104 RAFC-------------------
MMAR_5076|M.marinum_M RAFC-------------------
MUL_4152|M.ulcerans_Agy99 RAFC-------------------
Mflv_1451|M.gilvum_PYR-GCK ASFS-------------------
Mvan_5327|M.vanbaalenii_PYR-1 APFS-------------------
TH_1751|M.thermoresistible__bu AAFT-------------------
MAB_0574c|M.abscessus_ATCC_199 APFV-------------------
MSMEG_3401|M.smegmatis_MC2_155 RSPTTPASATTRKPILTAALGGE
.