For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TASHSSDHEEILVIGAGELGGSVVAALIRRDDAPEVTVLLRPSGTPRHARLRDELAALGAKVEQADVATA TVSELSAVFSRYDTVVSCIGFAAGPGTQRKITEAALAARVSRYFPWQFGVDYDAIGHGSPQDLFDEQLDV RDMLRAQDDTEWVIVSTGMFTSFLFEPDFGVVDLAANTVNALGSWDTEVTVTTPEDIGVLTAEIIQTRPR IANQVVYVAGDTISYRELADIVEKVRGVPVTRNEWTVAYLLEELAGRPDDGLAKYRAAFAQGKGVAWPKS GTFNEARGIATTTAEEWARANLDAGPAR*
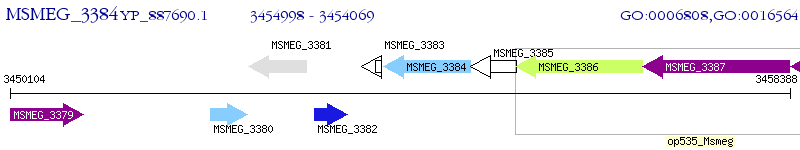
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3384 | - | - | 100% (309) | isoflavone reductase |
| M. smegmatis MC2 155 | MSMEG_6755 | - | e-104 | 64.21% (299) | isoflavone reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1263 | - | 2e-06 | 29.09% (165) | NAD dependent epimerase/dehydratase family protein |
| M. thermoresistible (build 8) | TH_3475 | - | 3e-06 | 24.78% (226) | PUTATIVE NAD dependent epimerase/dehydratase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1263|M.avium_104 -----------MIVGGHGKVALQLSAILTQRGDA--VTSLFR---NPDHA
TH_3475|M.thermoresistible__bu -------MARIVIIGGHGQVAMHLSRILSERGDQ--VTSVFR---NPNHA
MSMEG_3384|M.smegmatis_MC2_155 MTASHSSDHEEILVIGAGELGGSVVAALIRRDDAPEVTVLLRPSGTPRHA
::: * *::. : * .*.* ** ::* .* **
MAV_1263|M.avium_104 ---DDVAATGAKPVVADIERLDTDALAGLLAGHDAVVFSAGAGGGNPART
TH_3475|M.thermoresistible__bu ---EEVMVTGAQPLRADLEQLDTDTLTEVLSGHDAVVFSAGAGGGNPTRT
MSMEG_3384|M.smegmatis_MC2_155 RLRDELAALGAKVEQADVATATVSELSAVFSRYDTVVSCIGFAAGPGTQR
::: . **: **: .. *: ::: :*:** . * ..* ::
MAV_1263|M.avium_104 YAVDRDAAIRVVDAAARSGVKRFVMVSYFGAGPDHGVPQDDPFFPYAESK
TH_3475|M.thermoresistible__bu YAVDRDAAIRVIDAAGKAGVSRFIMVSYFGAGPDHGVPEDSSFFPYAESK
MSMEG_3384|M.smegmatis_MC2_155 KITEAALAARVSRYFPWQ-----FGVDYDAIG--HGSPQD----LFDEQL
.: * ** . *.* . * ** *:* : *.
MAV_1263|M.avium_104 AAADAHLRASDLDWTILGPGRLT---LDPPTGRIAVG--RGKGE------
TH_3475|M.thermoresistible__bu AAADAHLRASELDWTILGPGRLT---EEPATGKITVGPEAADSE------
MSMEG_3384|M.smegmatis_MC2_155 DVRDMLRAQDDTEWVIVSTGMFTSFLFEPDFGVVDLAANTVNALGSWDTE
. * .: :*.*:..* :* :* * : :. ..
MAV_1263|M.avium_104 ---VSRADVASVAAAALADDSTIRRTIDFNNGDVP-------IAVALAG-
TH_3475|M.thermoresistible__bu ---VPREDVALVAAACLADDSTVRRTIEFNKGDTS-------IAEAVRAR
MSMEG_3384|M.smegmatis_MC2_155 VTVTTPEDIGVLTAEIIQTRPRIANQVVYVAGDTISYRELADIVEKVRGV
.. *:. ::* : . : . : : **. *. : .
MAV_1263|M.avium_104 --------------------------------------------------
TH_3475|M.thermoresistible__bu --------------------------------------------------
MSMEG_3384|M.smegmatis_MC2_155 PVTRNEWTVAYLLEELAGRPDDGLAKYRAAFAQGKGVAWPKSGTFNEARG
MAV_1263|M.avium_104 --------------------
TH_3475|M.thermoresistible__bu --------------------
MSMEG_3384|M.smegmatis_MC2_155 IATTTAEEWARANLDAGPAR