For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PARTPRLRLKPKAPQSGYVDGAWWPHSDDLRSELPDLLAVLSVRLGRIDRVLYHLNAWAEAPRKLITGGR TVRLDGYRLQPINTIDVLGLDGDRLTLLVIPPHTNANDAHRTMMTAAQPHNAATVDGLLTISQRDRDTRN QASTAQERWESEGGTTASLRLAQLPG*
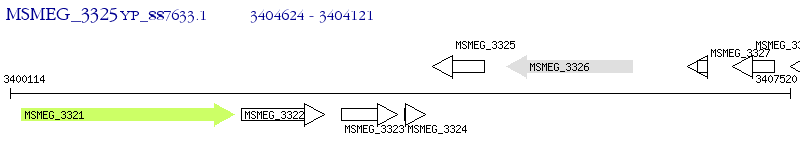
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3325 | - | - | 100% (167) | hypothetical protein MSMEG_3325 |
| M. smegmatis MC2 155 | MSMEG_3323 | - | 5e-49 | 62.09% (153) | hypothetical protein MSMEG_3323 |
| M. smegmatis MC2 155 | MSMEG_6678 | - | 5e-20 | 39.55% (134) | hypothetical protein MSMEG_6678 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5226 | - | 9e-38 | 59.54% (131) | hypothetical protein Mflv_5226 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2551 | - | 4e-05 | 31.97% (122) | hypothetical protein MAB_2551 |
| M. marinum M | MMAR_1227 | - | 4e-18 | 41.44% (111) | hypothetical protein MMAR_1227 |
| M. avium 104 | MAV_2295 | - | 8e-07 | 29.57% (115) | hypothetical protein MAV_2295 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2527 | - | 9e-15 | 40.38% (104) | hypothetical protein Mvan_2527 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3325|M.smegmatis_MC2_155 -----------------MPARTPRLRLKPKAPQSGYVDGAWWPHSDDLRS
Mflv_5226|M.gilvum_PYR-GCK MTQSGQDRGVGFGQPHNGPQSTPRLRLKPKAATTGFVDGAWWPHSTDLLA
MMAR_1227|M.marinum_M ------------------MERETRLQLRSFRPALEHVDGAWWPRSRQLVD
Mvan_2527|M.vanbaalenii_PYR-1 MANTPPPSSNSVGTTPVDVPMSGRLRLTRGRAPSEHIDGAWWPTSKRLDD
MAV_2295|M.avium_104 ------------MGSAARRRRADPVRLSVGCELGRAIDGAWWPRADRITN
MAB_2551|M.abscessus_ATCC_1997 --------------MTPVERSRPSIRFTLGPRSTGRMNGAWWPQDDGLIA
::: ::***** :
MSMEG_3325|M.smegmatis_MC2_155 -ELPDLLAVLSVRLGRIDRVLYHLNAWAEA-PRKLITGGRTVRLDGYRLQ
Mflv_5226|M.gilvum_PYR-GCK -ELPDLLAVLSVRLGDIARVAYNRGEWTSATPRKMLIDNRVVRLDGYDRQ
MMAR_1227|M.marinum_M -ELPGLVASITERLGPIVMVGYHRNGWEQT-PTVAEIAGHTVELLGFSSD
Mvan_2527|M.vanbaalenii_PYR-1 -QLPDLVAALRDRIPSVALVGYRRDGWIAP-PQVDLGDGHIVQLLSFASD
MAV_2295|M.avium_104 -ELPELVAVLTPLLGQIGAINVNWPPLQRP---------PDFNWPGWERK
MAB_2551|M.abscessus_ATCC_1997 RELSPLIEELSGRIGTVREVSLNWKAGSPR-----SSMRSVVMPVHLTSR
:*. *: : : : : . .
MSMEG_3325|M.smegmatis_MC2_155 PINTIDVLGLDGDRLTLLVIPPHTNANDAHRTMMTAAQPHNAATVDGLLT
Mflv_5226|M.gilvum_PYR-GCK PAHTIGVTGRGGWSNVLLVVPAATDSDDAHHVMMAAAAPGDRSAVDLLLT
MMAR_1227|M.marinum_M DPASLILIGQDGHHLTLRVISPDASDQSAQRTLDEVRVDTDAGSPPAGRS
Mvan_2527|M.vanbaalenii_PYR-1 EPPTIIVIGEDGHHLTLRVIDPDTDRHDAQVALDEIPRRSDSQRLSR---
MAV_2295|M.avium_104 RQHVMTLIG-GDARINLLIIPYATYSALARMVLRCAAGLPVEPRDRGKPA
MAB_2551|M.abscessus_ATCC_1997 PFHWVITLRGERRTVRMLLVPARTHRMLAMMVMRLAAQMPIVESPKE---
: : :: : * .:
MSMEG_3325|M.smegmatis_MC2_155 ISQRDRDTRNQASTAQERWESEGGTTASLRLAQLPG--------------
Mflv_5226|M.gilvum_PYR-GCK T----------------------GATG-----------------------
MMAR_1227|M.marinum_M LVERNVADVADKLARHEGLDDDQRTAQIKRWCEEAAAQFVDAPVQTFVPI
Mvan_2527|M.vanbaalenii_PYR-1 PAARSVAEVAKKLADHEGLNQVERNAELLRWCEDAATQFDEARIQTFVPI
MAV_2295|M.avium_104 FLTAG---------------SILRAAEQQRVDSAP---------------
MAB_2551|M.abscessus_ATCC_1997 ----------------------DEVTAALRIVMAA---------------
:
MSMEG_3325|M.smegmatis_MC2_155 --------------------------------
Mflv_5226|M.gilvum_PYR-GCK --------------------------------
MMAR_1227|M.marinum_M LVEHIVRNRMIESRGATRSPLLERSPEPANSR
Mvan_2527|M.vanbaalenii_PYR-1 LVEHIVNNRIHRERRAARDRTHDTEAPQTTS-
MAV_2295|M.avium_104 --------------------------------
MAB_2551|M.abscessus_ATCC_1997 --------------------------------