For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLHADAAHPDLVTVEVPTHWYNLAAELDQPIPPHLHPATKEPVGPDDLAALFPSGLIAQEVSTEAYIAI PDPVREIYSMWRPSPLIRARRFEKALNTGAHIYVKYEGVSPVGSHKTNSAVAQAYYNSVDGVRKLTTETG AGQWGSALAFAGAQFGLEIEVWQVRASYESKPYRGHLIRTYGGVVHSSPSELTESGRAILAKDPNTTGSL GMAVSEAVEVAAGDPDTRYALGSVLNHVVLHQSVIGQEAVAQLAAVEPNGADLVFGCAGGGSNLAGLAFP FLREKIHGRSDPKVIAAEPAACPSITQGEYRYDHGDVAGLTPLLKMHTLGMDFVPDPIHAGGLRYHGMAP ALSHTVELGLVQGVAISQHDAFSAGVQFARTQGIVPAPESTHAIAAAAAHVADDPSEQVVVIGLSGHGQL DLPAYAEYLDGKF
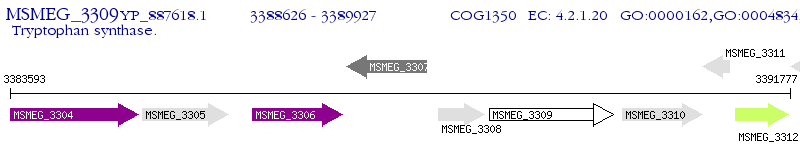
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3309 | - | - | 100% (433) | tryptophan synthase subunit beta |
| M. smegmatis MC2 155 | MSMEG_3220 | trpB | 8e-25 | 29.12% (364) | tryptophan synthase subunit beta |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1638 | trpB | 2e-23 | 28.53% (368) | tryptophan synthase subunit beta |
| M. gilvum PYR-GCK | Mflv_3175 | - | 0.0 | 89.61% (433) | tryptophan synthase subunit beta |
| M. tuberculosis H37Rv | Rv1612 | trpB | 2e-23 | 28.53% (368) | tryptophan synthase subunit beta |
| M. leprae Br4923 | MLBr_01272 | trpB | 1e-24 | 26.84% (395) | tryptophan synthase subunit beta |
| M. abscessus ATCC 19977 | MAB_2644c | - | 4e-26 | 29.51% (366) | tryptophan synthase subunit beta |
| M. marinum M | MMAR_2414 | trpB | 2e-24 | 27.76% (407) | tryptophan synthase, beta subunit TrpB |
| M. avium 104 | MAV_3174 | trpB | 4e-28 | 30.22% (364) | tryptophan synthase subunit beta |
| M. thermoresistible (build 8) | TH_0141 | - | 0.0 | 91.69% (433) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1592 | trpB | 3e-24 | 27.76% (407) | tryptophan synthase subunit beta |
| M. vanbaalenii PYR-1 | Mvan_2923 | - | 0.0 | 90.30% (433) | tryptophan synthase subunit beta |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1638|M.bovis_AF2122/97 ---------------MSAAIAEPTSHDPDSGGHFGG---PSGWGGRYVPE
Rv1612|M.tuberculosis_H37Rv ---------------MSAAIAEPTSHDPDSGGHFGG---PSGWGGRYVPE
MMAR_2414|M.marinum_M ----MADLSRPDLPRTSSAIAERTSHDPDSGGHFG------AYGGRYVPE
MUL_1592|M.ulcerans_Agy99 ----MADLSRPDLPRTSSAIAERTSHDPDSGGHFG------AYGGRYVPE
MLBr_01272|M.leprae_Br4923 --------MMPNLSCFSVAISEPTCHDPDSGGHFGG---PDGYGGRYVPE
MAV_3174|M.avium_104 MTVDISPRTRPDLPLPSAAIAEPTRHEPDAGGHFG------VYGGRYVAE
MAB_2644c|M.abscessus_ATCC_199 -------MKNTGLPQMSTAIAEPTAHDPDARGHFG------PYGGRYVAE
MSMEG_3309|M.smegmatis_MC2_155 ------MTLHADAAHPDLVTVEVPTHWYNLAAELDQPIPPHLHPATKEPV
TH_0141|M.thermoresistible__bu ------MTQQADATHPDLVTVGVPTHWYNLAAELDEPIPPHLHPATREPV
Mflv_3175|M.gilvum_PYR-GCK ------MTEQRGATHPDLVAAEVPTHWYNLAAELDEPIPPHLHPGTKEPV
Mvan_2923|M.vanbaalenii_PYR-1 ------MTEQLDATHPDLVTVKVPTHWYNLAAELDEPIPPHLHPGTREPV
. . . * : ..:. . .
Mb1638|M.bovis_AF2122/97 ALMAVIEEVTAAYQKERVS-QDFLDDLDRLQANYAG-RPSPLYEATRLSQ
Rv1612|M.tuberculosis_H37Rv ALMAVIEEVTAAYQKERVS-QDFLDDLDRLQANYAG-RPSPLYEATRLSQ
MMAR_2414|M.marinum_M ALMAVIEEVTVAYEKERVN-QDFLDTLDRLQTHYAG-RPSPLYEATRLSE
MUL_1592|M.ulcerans_Agy99 ALMAVIEEVAVAYEKERVN-QDFLDTLDRLQTHYAG-RPSPLYEATRLSA
MLBr_01272|M.leprae_Br4923 ALMAVIEEVTAAYEKERVN-QDFLDLLDKLQANYAG-RPSPLYEATRLSE
MAV_3174|M.avium_104 ALMAVIEEVTTAYEKERVN-QDFLDTLDYLQANYAG-RPSPLYEAPRLSE
MAB_2644c|M.abscessus_ATCC_199 ALMAVIEEVTAAYEKIRSD-RSFLDELDRLQTHYVG-RPSPLYEAERLSA
MSMEG_3309|M.smegmatis_MC2_155 GPDDLAALFPSGLIAQEVSTEAYIAIPDPVREIYSMWRPSPLIRARRFEK
TH_0141|M.thermoresistible__bu GPDDLAPLFASGLIAQEVSTETYIAIPEPVREIYAMWRPSPLIRARRFEQ
Mflv_3175|M.gilvum_PYR-GCK SPDDLAPLFASGLIAQEVATETYIEIPEPVREILSMWRPSPLIRARRFEK
Mvan_2923|M.vanbaalenii_PYR-1 GPDDLAPLFASALIAQEVASETYIEIPQEVREIYAMWRPSPLIRARRFEK
. : .. . . . :: : :: ***** .* *:.
Mb1638|M.bovis_AF2122/97 HAGS-ARIFLKREDLNHTGSHKINNVLGQALLARRMGKTRVIAETGAGQH
Rv1612|M.tuberculosis_H37Rv HAGS-ARIFLKREDLNHTGSHKINNVLGQALLARRMGKTRVIAETGAGQH
MMAR_2414|M.marinum_M HAGS-ARIFLKREDLNHTGSHKINNVLGQALLARRMGKTRVIAETGAGQH
MUL_1592|M.ulcerans_Agy99 HAGS-ARIFLKREDLNHTGSHKINNVLGQALLARRMGKTRVIAETGAGQH
MLBr_01272|M.leprae_Br4923 YAGS-VRIFLKREDLNHTGSHKINNVLGQTLLAQRMGKTRVIAETGAGQH
MAV_3174|M.avium_104 QAG--ARIFLKREDLNHTGSHKINNVLGQALLAQRMGKKRVIAETGAGQH
MAB_2644c|M.abscessus_ATCC_199 HAGG-ARIFLKREDLNHTGSHKINNVLGQVLLAKKMGKTRVIAETGAGQH
MSMEG_3309|M.smegmatis_MC2_155 ALNTGAHIYVKYEGVSPVGSHKTNSAVAQAYYNSVDGVRKLTTETGAGQW
TH_0141|M.thermoresistible__bu ALNTGARIYVKYEGVSPVGSHKTNSAVAQAYYNSIDGVRKLTTETGAGQW
Mflv_3175|M.gilvum_PYR-GCK ALNTGARIYVKYEGVSPVGSHKTNSAVAQAYYNSLDGVRKLTTETGAGQW
Mvan_2923|M.vanbaalenii_PYR-1 ALNTGAHIYVKYEGVSPVGSHKTNSAVAQAYYNHIDGVRKLTTETGAGQW
. .:*::* *.:. .**** *..:.*. * :: :******
Mb1638|M.bovis_AF2122/97 GVATATACALLGLDCVIYMGGIDTARQALNVARMRLLGAEVVA-----VQ
Rv1612|M.tuberculosis_H37Rv GVATATACALLGLDCVIYMGGIDTARQALNVARMRLLGAEVVA-----VQ
MMAR_2414|M.marinum_M GVATATACALLGLECVIYMGAVDTARQALNVARMRLLGAEVVA-----VE
MUL_1592|M.ulcerans_Agy99 GVATATACALLGLECVIYMGAVDTARQALNVARMRLLGAEVVA-----VE
MLBr_01272|M.leprae_Br4923 GVATATACALFGLDCVIYMGALDTARQALNVARMRLLGAEVVS-----VE
MAV_3174|M.avium_104 GVATATACALLGLECVIYMGAVDTERQALNVARMRLLGATVVS-----VQ
MAB_2644c|M.abscessus_ATCC_199 GVATATACALLGLECVIYMGAVDTARQALNVARMRLLGSTVVS-----VE
MSMEG_3309|M.smegmatis_MC2_155 GSALAFAGAQFGLEIEVWQVRASYESKPYRGHLIRTYGGVVHSSPSELTE
TH_0141|M.thermoresistible__bu GSALAFAGAQFGLDIEVWQVRASYDSKPYRGHLIRTYGGTVHPSPSDLTE
Mflv_3175|M.gilvum_PYR-GCK GSALSFAGAQFGLEIEVWQVRASYESKPYRGHLMRTYGGTVHPSPSTLTE
Mvan_2923|M.vanbaalenii_PYR-1 GSALSFAGAQFGLEVEVWQVRASYESKPYRGHLIRTYGGTVHSSPSTLTE
* * : * * :**: :: . :. . :* *. * . .:
Mb1638|M.bovis_AF2122/97 TGSKTLKDAINEAFRDWVANADNTYYCFGTAAGPHPFPTMVR---DFQRI
Rv1612|M.tuberculosis_H37Rv TGSKTLKDAINEAFRDWVANADNTYYCFGTAAGPHPFPTMVR---DFQRI
MMAR_2414|M.marinum_M TGSKTLKDAINEAFRDWVTNADRTYYCFGTAAGPHPFPTMVR---DFQRI
MUL_1592|M.ulcerans_Agy99 TGSKTLKDAINEAFRDWVTNADRTYYCFGTAAGPHPFPTMVR---DFQRI
MLBr_01272|M.leprae_Br4923 TGSRTLKDAINDAFRDWVTNADNTYYCFGTASGPHPFPTMVR---DLQRV
MAV_3174|M.avium_104 SGSKTLKDAINEAFRDWVTNADNTFYCFGTAAGPHPFPAMVR---DFQRI
MAB_2644c|M.abscessus_ATCC_199 SGSKTLKDAINDAMRDWVTNAHNTYYCFGTAAGPHPFPVMVR---DFQRI
MSMEG_3309|M.smegmatis_MC2_155 SGRAILAKDPNTTGSLGMAVSEAVEVAAGDPDTRYALGSVLNHVVLHQSV
TH_0141|M.thermoresistible__bu AGRAILASDPNTTGSLGMAVSEAVEVAASEPDTRYALGSVLNHVVLHQSV
Mflv_3175|M.gilvum_PYR-GCK SGRAILAKDPDTTGSLGMAVSEAVEVAAADPEARYALGSVLNHVVLHQTV
Mvan_2923|M.vanbaalenii_PYR-1 AGRAILAKDPDTTGSLGMAVSEAVEVAAADPEARYALGSVLNHVVLHQTV
:* * . : : :: :. . . . . :.: ::. * :
Mb1638|M.bovis_AF2122/97 IGMEARVQIQGQAGRLPDAVVACVGGGSNAIGIFHAFLDDPGVRLVG-FE
Rv1612|M.tuberculosis_H37Rv IGMEARVQIQGQAGRLPDAVVACVGGGSNAIGIFHAFLDDPGVRLVG-FE
MMAR_2414|M.marinum_M IGMEARVQIQGQAGKLPDAVVACVGGGSNAIGIFHAFLDDPDVRLVG-FE
MUL_1592|M.ulcerans_Agy99 IGMEARVQIQGQAGKLPDAVVACVGGGSNAIGIFHAFLDDPDVRLVG-FE
MLBr_01272|M.leprae_Br4923 IGLETRRQIQYQAGRLPDAVTACIGGGSNAIGIFHAFLDDPGVRLVG-FE
MAV_3174|M.avium_104 IGLEARAQIQAQAGRLPDAVVACIGGGSNAIGIFHPFIDDPGVRLVG-FE
MAB_2644c|M.abscessus_ATCC_199 IGMEARAQVLDQVGRLPDAAVACVGGGSNAIGLFHAFIDDPAVRLVG-YE
MSMEG_3309|M.smegmatis_MC2_155 IGQEAVAQLAAVEPNGADLVFGCAGGGSNLAGLAFPFLREKIHGRSDPKV
TH_0141|M.thermoresistible__bu IGQEAVDQLALVEPGGADAVFGCAGGGSNLAGLAFPFLREKIHGRSNPRV
Mflv_3175|M.gilvum_PYR-GCK IGQEAVAQLALTEPNGADVVFGCAGGGSNLAGLSFPFLREKIHGRSNPRI
Mvan_2923|M.vanbaalenii_PYR-1 IGQEAVAQLALVEPDGADVVFGCAGGGSNLAGLSFPFLREKIHGRSNPRI
** *: *: .* . .* ***** *: ..*: : .
Mb1638|M.bovis_AF2122/97 AAGDGVETGRHAATFTAGSPGAFHGSFSYLLQDEDGQTIESHSISAG-LD
Rv1612|M.tuberculosis_H37Rv AAGDGVETGRHAATFTAGSPGAFHGSFSYLLQDEDGQTIESHSISAG-LD
MMAR_2414|M.marinum_M AAGDGVETGRHAATFAGGSPGAFQGSFSYLLQDEDGQTIESHSISAG-LD
MUL_1592|M.ulcerans_Agy99 AAGDGVETGRHAATFAGGSPGAFQGSFSYLLQDEDGQTIESHSISAG-LD
MLBr_01272|M.leprae_Br4923 AAGDGVETGRHAATLTGGLPGAFQGTFSYLLQDEDGQTIESHSIAAG-LD
MAV_3174|M.avium_104 AAGDGVETGRHAATFSGGSPGAFQGSFSYLLQDEDGQTIESHSISAG-LD
MAB_2644c|M.abscessus_ATCC_199 AAGDGVETGRHAATFTGGTPGAFQGSYSYLLQDDDGQTIESHSISAG-LD
MSMEG_3309|M.smegmatis_MC2_155 IAAEPAACPSITQGEYRYDHGDVAGLTPLLKMHTLGMDFVPDPIHAGGLR
TH_0141|M.thermoresistible__bu VAAEPAACPSITQGEYRYDHGDVAGLTPLLKMHTLGMDFVPDPIHAGGLR
Mflv_3175|M.gilvum_PYR-GCK VAAEPSACPSITQGEYRYDHGDVAGLTPLLKMHTLGMDFVPDSIHAGGLR
Mvan_2923|M.vanbaalenii_PYR-1 VAAEPAACPSITRGEYRYDHGDVAGLTPLLKMHTLGMDFVPDPIHAGGLR
*.: : * . * . * . * : ...* ** *
Mb1638|M.bovis_AF2122/97 YPGVGPEHAWLKEAGRVDYRPITDSEAMDAFGLLCRMEGIIPAIESAHAV
Rv1612|M.tuberculosis_H37Rv YPGVGPEHAWLKEAGRVDYRPITDSEAMDAFGLLCRMEGIIPAIESAHAV
MMAR_2414|M.marinum_M YPGVGPEHAWLKDIGRVTYQPITDSEAMEAFGLLCRMEGIIPAIESAHAV
MUL_1592|M.ulcerans_Agy99 YPGVGPEHAWLKDIGRVTYQPITDSEAMEAFGLLCRMEGIIPAIESAHAV
MLBr_01272|M.leprae_Br4923 YPGVGPEHAWLRETGRAEYQPITDSEAIEAFRLLCRTEGILPALESAHAV
MAV_3174|M.avium_104 YPGVGPEHAYLRETGRAEYRPITDAEAMDAFRVLCRTEGIIPAIESAHAV
MAB_2644c|M.abscessus_ATCC_199 YPGVGPEHAWLRESGRADYRPITDGEAMDAFLLLSRAEGIIPAIESAHAV
MSMEG_3309|M.smegmatis_MC2_155 YHGMAPALSHTVELGLVQGVAISQHDAFSAGVQFARTQGIVPAPESTHAI
TH_0141|M.thermoresistible__bu YHGMAPALSHTVELGLVEAVAISQHDAFAAGVQFARSQGIVPAPESTHAI
Mflv_3175|M.gilvum_PYR-GCK YHGMAPALSHTVELGLVEGVAISQHDAFAAGVQFARTQGIVPAPESTHAI
Mvan_2923|M.vanbaalenii_PYR-1 YHGMAPALSHTVELGLVEGVAISQHDAFAAGVQFARTQGIVPAPESTHAI
* *:.* : : * . .*:: :*: * :.* :**:** **:**:
Mb1638|M.bovis_AF2122/97 AGALKLGVELGRGAVIVVNLSGRGDKDVETAAKWFGLLGND-------
Rv1612|M.tuberculosis_H37Rv AGALKLGVELGRGAVIVVNLSGRGDKDVETAAKWFGLLGND-------
MMAR_2414|M.marinum_M AGALKLGRELGKGAVIVVNLSGRGDKDVETAAKWFGLLTPGGSANKSV
MUL_1592|M.ulcerans_Agy99 AGALKLGRELGKGAVIVVNLSGRGDKDVETAAKWFGLLTPGGSANKSV
MLBr_01272|M.leprae_Br4923 AGALKLASEIGKGAVIVVNLSGRGDKDIEAAAKWFGLLGTR-------
MAV_3174|M.avium_104 AGALKLGPELGRGAIIVVNLSGRGDKDVETAAKWFGLLGSSDR-----
MAB_2644c|M.abscessus_ATCC_199 AGALKLGGELSEGAVIVVSLSGRGDKDVETAAKWFDLLEKDGSQS---
MSMEG_3309|M.smegmatis_MC2_155 AAAAAHVADDPSEQVVVIGLSGHGQLDLPAYAEYLDGKF---------
TH_0141|M.thermoresistible__bu AAAAAQVADDPREQVVVIGLSGHGQLDLPAYAEYLDGKF---------
Mflv_3175|M.gilvum_PYR-GCK AAAARYVADNPREQVVVIGLSGHGQLDLPAYAEYLDGKF---------
Mvan_2923|M.vanbaalenii_PYR-1 AAAAAHVADDPREQVVVIGLSGHGQLDLPAYAEYLDGKF---------
*.* : ::*:.***:*: *: : *:::.