For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSRRGKIVCTLGPATSTDETVRALVEAGMDVARLNFSHGDYSDHEAAYKRVRAASDVTGHAVGIMADLQG PKIRLGRFKDGPTYWATGETVRITVEDCEGTHDRVSTTYKQLAQDAARGDRLLVDDGKVALKVEEVDGDD VVCTVTEGGPVSNNKGVSLPGMNVSVPAMSEKDIEDLEFAVRLGVDLVALSFVRSPADIELVHEVMDRVG RRVPVIAKLEKPEAIENLEAVVLAFDAVMVARGDLGVELPLEEVPLVQKRAIQMARENAKPVIVATQMLE SMIENSRPTRAEASDVANAVLDGADAVMLSGETSVGKYPLEAVRTMARIIQAVEDNSVVVPPLTHVPRTK RGVISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLPVLAFTALPEVRSQLALTWGTETFIVPQMS NTDDMIRQVDKSLLELGRYKRGELVVIVAGAPPGTVGSTNLIHVHRIGEDDV
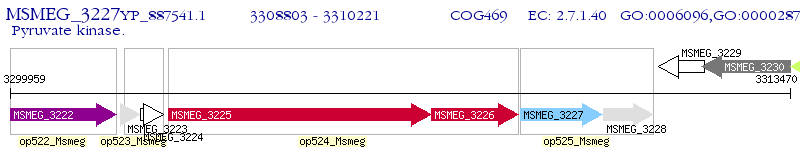
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3227 | pyk | - | 100% (472) | pyruvate kinase |
| M. smegmatis MC2 155 | MSMEG_0154 | pyk | e-149 | 56.54% (474) | pyruvate kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1643 | pykA | 0.0 | 86.23% (472) | pyruvate kinase |
| M. gilvum PYR-GCK | Mflv_3591 | - | 0.0 | 87.92% (472) | pyruvate kinase |
| M. tuberculosis H37Rv | Rv1617 | pykA | 0.0 | 86.44% (472) | pyruvate kinase |
| M. leprae Br4923 | MLBr_01277 | pykA | 0.0 | 85.17% (472) | pyruvate kinase |
| M. abscessus ATCC 19977 | MAB_2639c | - | 0.0 | 85.14% (471) | pyruvate kinase |
| M. marinum M | MMAR_2420 | pykA | 0.0 | 86.65% (472) | pyruvate kinase PykA |
| M. avium 104 | MAV_3170 | pyk | 0.0 | 88.44% (450) | pyruvate kinase |
| M. thermoresistible (build 8) | TH_1097 | pykA | 0.0 | 87.81% (443) | Probable pyruvate kinase pykA |
| M. ulcerans Agy99 | MUL_1598 | pykA | 0.0 | 86.65% (472) | pyruvate kinase |
| M. vanbaalenii PYR-1 | Mvan_2824 | - | 0.0 | 88.77% (472) | pyruvate kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3591|M.gilvum_PYR-GCK MTRRGKIVCTLGPATASDEAVRQLVESGMDVARLNFSHGDYPDHEANYKR
Mvan_2824|M.vanbaalenii_PYR-1 MNRRGKIVCTLGPATATEEAVRQLVESGMDVARLNFSHGDYPDHEANYKR
MSMEG_3227|M.smegmatis_MC2_155 MSRRGKIVCTLGPATSTDETVRALVEAGMDVARLNFSHGDYSDHEAAYKR
Mb1643|M.bovis_AF2122/97 MTRRGKIVCTLGPATQRDDLVRALVEAGMDVARMNFSHGDYDDHKVAYER
Rv1617|M.tuberculosis_H37Rv MTRRGKIVCTLGPATQRDDLVRALVEAGMDVARMNFSHGDYDDHKVAYER
MMAR_2420|M.marinum_M MTRRGKIVCTLGPATQQDDLVKELVEAGMDVARMNFSHGDYPDHEAAYER
MUL_1598|M.ulcerans_Agy99 MTRRGKIVCTLGPATQQDDLVKELVEAGMDVARMNFSHGDYADHEAAYER
MLBr_01277|M.leprae_Br4923 MTRRGKIVCTLGPATQSDELIRALVEAGMDVARMNFSHGDYSDHKAVYDR
MAV_3170|M.avium_104 ---------------------MALVEAGMDVARLNFSHGDYADHKAAYER
TH_1097|M.thermoresistible__bu ----------------------------MDVARLNFSHGTHADHERYYKL
MAB_2639c|M.abscessus_ATCC_199 MTRRGKIVCTLGPATGSAELVRALVEAGMDVARLNFSHGEHADHEQNYRW
*****:***** : **: *
Mflv_3591|M.gilvum_PYR-GCK VRAASDATGRAVGILADLQGPKIRLGRFADGPTVWRNGETIRITVDDFEG
Mvan_2824|M.vanbaalenii_PYR-1 VRAASDATGRAVGILADLQGPKIRLGRFADGPTMWANGETVRITVEDFVG
MSMEG_3227|M.smegmatis_MC2_155 VRAASDVTGHAVGIMADLQGPKIRLGRFKDGPTYWATGETVRITVEDCEG
Mb1643|M.bovis_AF2122/97 VRVASDATGRAVGVLADLQGPKIRLGRFASGATHWAEGETVRITVGACEG
Rv1617|M.tuberculosis_H37Rv VRVASDATGRAVGVLADLQGPKIRLGRFASGATHWAEGETVRITVGACEG
MMAR_2420|M.marinum_M VRAASDATGRAVGVLADLQGPKIRLGRFADGPTYWADGDTVRITVEECEG
MUL_1598|M.ulcerans_Agy99 VRAASDATGRAVGVLADLQGPKIRLGRFADGPTYWADGDTVRITVEECEG
MLBr_01277|M.leprae_Br4923 VRVASDATGRAVGVLADLQGPKIRLGRFATGSTYWADGETVRITVADCHG
MAV_3170|M.avium_104 VRVASDATGRAVGVLADLQGPKIRLGRFATGPTYWADGETVRITVADCEG
TH_1097|M.thermoresistible__bu VRAASDLTGRAVGILADLQGPKIRLGRFADGSTYWATGETVRITVDDIQG
MAB_2639c|M.abscessus_ATCC_199 VRKASDESGRAVGVLADLQGPKIRLGRFATGSAHWATGEQVRITVEDVVG
** *** :*:***::************* *.: * *: :**** *
Mflv_3591|M.gilvum_PYR-GCK THDRVSTTYKRLAQDATPGDRVLVDDGNVGLVVEHIDGNDVVCTVTEGGK
Mvan_2824|M.vanbaalenii_PYR-1 NHDRVSTTYKRLAQDAKPGDRVLVDDGNVGLVVEHIDGNDVVCTVTEGGK
MSMEG_3227|M.smegmatis_MC2_155 THDRVSTTYKQLAQDAARGDRLLVDDGKVALKVEEVDGDDVVCTVTEGGP
Mb1643|M.bovis_AF2122/97 SHDRVSTTYKRLAQDAVAGDRVLVDDGKVALVVDAVEGDDVVCTVVEGGP
Rv1617|M.tuberculosis_H37Rv SHDRVSTTYKRLAQDAVAGDRVLVDDGKVALVVDAVEGDDVVCTVVEGGP
MMAR_2420|M.marinum_M CRDRVSTTYKRLAQDAVAGDRVLVDDGKVGLIVDHIDGNDVVCTVVEGGP
MUL_1598|M.ulcerans_Agy99 CRDRVSTTYKRLAQDAVAGDRVLVDDGKVGLIVDHIDGNDVVCTVVEGGP
MLBr_01277|M.leprae_Br4923 SHDRVSTTYKNLARDAVVGDRVLVDDGKVGLVVSAIDGDDVVCTVTEGGP
MAV_3170|M.avium_104 SHDRVSTTYKQLAQDAAVGDRVLVDDGKVGLVVDGIEGDDVICTVVEGGP
TH_1097|M.thermoresistible__bu THDRVSTTYKNLAADAARGDRLLVDDGKVALVVTAVEGDDVVCEVVEGGP
MAB_2639c|M.abscessus_ATCC_199 TPDRVSTTYKQLAADAAVGDRLLVDDGKVGLTVEAIEGDDVVCRVTEGGP
********.** ** ***:*****:*.* * ::*:**:* *.***
Mflv_3591|M.gilvum_PYR-GCK VSNNKGMSLPGMNVSAPALSEKDIDDLEFALRLGVDLVALSFVRSPADVE
Mvan_2824|M.vanbaalenii_PYR-1 VSNNKGMSLPGMNVSAPALSEKDIDDLEFALRLGVDLVALSFVRSPADVE
MSMEG_3227|M.smegmatis_MC2_155 VSNNKGVSLPGMNVSVPAMSEKDIEDLEFAVRLGVDLVALSFVRSPADIE
Mb1643|M.bovis_AF2122/97 VSDNKGISLPGMNVTAPALSEKDIEDLTFALNLGVDMVALSFVRSPADVE
Rv1617|M.tuberculosis_H37Rv VSDNKGISLPGMNVTAPALSEKDIEDLTFALNLGVDMVALSFVRSPADVE
MMAR_2420|M.marinum_M VSNNKGISLPGMNVSAPALSEKDIDDLTFALNLGVDMVALSFVRSPSDVE
MUL_1598|M.ulcerans_Agy99 VSNNKGISLPGMNVSAPALSEKDIDDLTFALNLGVDMVALSFVRSPSDVE
MLBr_01277|M.leprae_Br4923 VSNNKGMSLPGMHVSAPALSDKDIEDLTFALNLGADLVALSFVRSPSDVE
MAV_3170|M.avium_104 VSNNKGISLPGMNVSAPALSDKDIEDLTFALELGVDLVALSFVRSPSDVE
TH_1097|M.thermoresistible__bu VSNNKGLSLPGMNVSVPALTEKDVEDLEFAIRLGVDMVALSFVRSPADVE
MAB_2639c|M.abscessus_ATCC_199 VSNNKGLSLPGMNVSVPALSEKDIADLEFALRLGVDLIALSFVRSPSDVE
**:***:*****:*:.**:::**: ** **:.**.*::********:*:*
Mflv_3591|M.gilvum_PYR-GCK LVHEIMDRVGRRVPVIAKLEKPEAIDNLEAIVLAFDAIMVARGDLGVELP
Mvan_2824|M.vanbaalenii_PYR-1 LVHEVMDRVGRRVPVIAKLEKPEAIDNLEAIVLAFDAIMVARGDLGVELP
MSMEG_3227|M.smegmatis_MC2_155 LVHEVMDRVGRRVPVIAKLEKPEAIENLEAVVLAFDAVMVARGDLGVELP
Mb1643|M.bovis_AF2122/97 LVHEVMDRIGRRVPVIAKLDKPEAIDNLEAIVLAFDAVMVARGDLGVELP
Rv1617|M.tuberculosis_H37Rv LVHEVMDRIGRRVPVIAKLEKPEAIDNLEAIVLAFDAVMVARGDLGVELP
MMAR_2420|M.marinum_M LVHEVMDRIGRRVPVIAKLEKPEAIDNLEAIVLAFDAVMVARGDLGVELP
MUL_1598|M.ulcerans_Agy99 LVHEVMDRIGRRVPVIAKLEKPEAIDNLEAIVLAFDAVMVARGDLGVELP
MLBr_01277|M.leprae_Br4923 LVHKAMDRVGRRVPVIAKLEKPEAVDNLEAIVLAFDAIMVARGDLGVELP
MAV_3170|M.avium_104 LVHEVMDRVGRRVPVIAKLEKPEAVDNLEAIVLAFDAIMVARGDLGVELP
TH_1097|M.thermoresistible__bu LVHEVMDRVGRRVPVIAKLEKPEAIDNLEAIVLAFDGVMVARGDLGVELP
MAB_2639c|M.abscessus_ATCC_199 LVHAVMDRVGRRVPVIAKLEKPEAVANLEAIVLAFDAIMVARGDLGVELP
*** ***:**********:****: ****:*****.:************
Mflv_3591|M.gilvum_PYR-GCK LEEVPLVQKRAIQMARENAKPVIVATQMLESMIENSRPTRAEASDVANAV
Mvan_2824|M.vanbaalenii_PYR-1 LEEVPLVQKRAIQMARENAKPVIVATQMLESMIENSRPTRAEASDVANAV
MSMEG_3227|M.smegmatis_MC2_155 LEEVPLVQKRAIQMARENAKPVIVATQMLESMIENSRPTRAEASDVANAV
Mb1643|M.bovis_AF2122/97 LEEVPLVQKRAIQMARENAKPVIVATQMLDSMIENSRPTRAEASDVANAV
Rv1617|M.tuberculosis_H37Rv LEEVPLVQKRAIQMARENAKPVIVATQMLDSMIENSRPTRAEASDVANAV
MMAR_2420|M.marinum_M LEEVPLVQKRAIQIARENAKPVIVATQMLDSMIENSRPTRAEASDVANAV
MUL_1598|M.ulcerans_Agy99 LEEVPLVQKRAIQIARENAKPVIVATQMLDSMIENSRPTRAEASDVANAV
MLBr_01277|M.leprae_Br4923 LEEVPLVQKRAIQMARENAKPVIVATQMLDSMIENSRPTRAEASDVANAV
MAV_3170|M.avium_104 LEEVPLVQKRAIQMARENAKPVIVATQMLDSMIENSRPTRAEASDVANAV
TH_1097|M.thermoresistible__bu LEEVPLVQKRAIQTARENAKPVIVATQMLESMIENSRPTRAEASDVANAV
MAB_2639c|M.abscessus_ATCC_199 LEDVPIVQKRAIQIARENARPVIVATQMLESMIENSRPTRAEASDVANAV
**:**:******* *****:*********:********************
Mflv_3591|M.gilvum_PYR-GCK LDGADAVMLSGETSVGKFPFETVRTMARIVSAVEDNSVAVPALTHTPRTK
Mvan_2824|M.vanbaalenii_PYR-1 LDGADAVMLSGETSVGKFPFETVRTMARIIKAVEENSVAAPPLTHVPRTK
MSMEG_3227|M.smegmatis_MC2_155 LDGADAVMLSGETSVGKYPLEAVRTMARIIQAVEDNSVVVPPLTHVPRTK
Mb1643|M.bovis_AF2122/97 LDGADALMLSGETSVGKYPLAAVRTMSRIICAVEENSTAAPPLTHIPRTK
Rv1617|M.tuberculosis_H37Rv LDGADALMLSGETSVGKYPLAAVRTMSRIICAVEENSTAAPPLTHIPRTK
MMAR_2420|M.marinum_M LDGADALMLSGETSVGKYPLAAVRTMSRIVCAVEENSTAAPPLTHVPRTK
MUL_1598|M.ulcerans_Agy99 LDGADALMLSGETSVGKYPLAAVRTMSRIVCAVEENSTAAPPLTHVPRTK
MLBr_01277|M.leprae_Br4923 LDGADALMLSEETSVGKYPLAAVKTMSRIVCAVEENSTAAPPLTHVPRTK
MAV_3170|M.avium_104 LDGADAVMLSGETSVGKYPLAAVRTMARIICAVEDNSTAAPPLTHVPRTK
TH_1097|M.thermoresistible__bu LDGTDAVMLSGETSVGKYPLEAVRTMASIVAAVEENSTAAPPLTHLPRTK
MAB_2639c|M.abscessus_ATCC_199 LDGADAVMLSGETSVGKYPLEAVRTMARIVSAVESHSVSAPPLTHVPRTK
***:**:*** ******:*: :*:**: *: ***.:*. .*.*** ****
Mflv_3591|M.gilvum_PYR-GCK RGVISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLPVLAFTSLPE
Mvan_2824|M.vanbaalenii_PYR-1 RGVISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLPVLAFTALPE
MSMEG_3227|M.smegmatis_MC2_155 RGVISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLPVLAFTALPE
Mb1643|M.bovis_AF2122/97 RGVISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLPLLAFTAWPE
Rv1617|M.tuberculosis_H37Rv RGVISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLPLLAFTAWPE
MMAR_2420|M.marinum_M RGVISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLPLLAFTAWPE
MUL_1598|M.ulcerans_Agy99 RGVISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLPLLAFTAWPE
MLBr_01277|M.leprae_Br4923 RGVISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLPLLAFTAWPE
MAV_3170|M.avium_104 RGVISYAARDIGERLDAKALVAFTQSGDTVKRLARLHTPLPLLAFTAWPE
TH_1097|M.thermoresistible__bu RGVISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLPLLAFTSLPE
MAB_2639c|M.abscessus_ATCC_199 RGVISYAARDIGERLNAKALVAFTQSGDTVRRLARLHTPLPVLAFTPLPE
***************:**************:**********:****. **
Mflv_3591|M.gilvum_PYR-GCK VRSQLALTWGTETFIVPHIDTTDGMIRQVDQSMLNLGRYKRGDLVVIVAG
Mvan_2824|M.vanbaalenii_PYR-1 VRSQLALTWGTETFIVPHIDTTDGMIRQVDQSMLNLGRYKRGDLVVIVAG
MSMEG_3227|M.smegmatis_MC2_155 VRSQLALTWGTETFIVPQMSNTDDMIRQVDKSLLELGRYKRGELVVIVAG
Mb1643|M.bovis_AF2122/97 VRSQLAMTWGTETFIVPKMQSTDGMIRQVDKSLLELARYKRGDLVVIVAG
Rv1617|M.tuberculosis_H37Rv VRSQLAMTWGTETFIVPKMQSTDGMIRQVDKSLLELARYKRGDLVVIVAG
MMAR_2420|M.marinum_M VRSQLAMTWGTETFIVPEMDSTDGMIRQVDKSLLELGRYKRGDLVVIVAG
MUL_1598|M.ulcerans_Agy99 VRSQLAMTWGTETFIVPEMDSTDGMIRQVDKSLLELGRYKRGDLVVIVAG
MLBr_01277|M.leprae_Br4923 VRSQLAMTWGTETFLVPKMLSTDGMIRQVDKSLLELGRYKRGDLVVIVAG
MAV_3170|M.avium_104 VRSQLAMTWGTETFIVPMMTSTDGMIRQVDKSLLELGRYRRGDLVVIVAG
TH_1097|M.thermoresistible__bu IRSQLALTWGTETFIVPHIQTTDGMIRQVDQSLLQLGRYKRGDLVVIVAG
MAB_2639c|M.abscessus_ATCC_199 VRSQLALTWGTETFIVPVMETTDGMIQQVDTALLEMERYQRGDLVVIVAG
:*****:*******:** : .**.**:*** ::*:: **:**:*******
Mflv_3591|M.gilvum_PYR-GCK APPGTVGSTNLIHVHRIGEDDV-
Mvan_2824|M.vanbaalenii_PYR-1 APPGTVGSTNLIHVHRIGEDDV-
MSMEG_3227|M.smegmatis_MC2_155 APPGTVGSTNLIHVHRIGEDDV-
Mb1643|M.bovis_AF2122/97 APPGTVGSTNLIHVHRIGEDDV-
Rv1617|M.tuberculosis_H37Rv APPGTVGSTNLIHVHRIGEDDV-
MMAR_2420|M.marinum_M APPGTVGSTNLIHVHRIGEDDV-
MUL_1598|M.ulcerans_Agy99 APPGTVGSTNLIHVHRIGEDDV-
MLBr_01277|M.leprae_Br4923 TPPGTVGSTNLIHVHRIGEDDV-
MAV_3170|M.avium_104 APPGTVGSTNLIHVHRIGEDDV-
TH_1097|M.thermoresistible__bu APPGTVGSTNMIHVHRIGEDDHF
MAB_2639c|M.abscessus_ATCC_199 APPGTVGSTNLIHVHRIGEDDH-
:*********:**********