For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MANRRAKIVCTLGPATATSSVLTELVDAGMDVARLNFSHSTHAEHSALYGMVREIAAQRGRVVGVLADLQ GPKIRLGCFADGPVVWATGEHVTITTEDCPGDHDRVSTTYAGLSQDVRAGDRLLVDDGRVDLRVVAVDGP DIRCEVVDGGPVSDHKGISLPNIPVSVPPLSDKDIEDLKFALELGADMIAMSFVRAPEEVELAHKIMDEV GRRVPVIAKLEKPEAVSDLPAIVEAFDGLMVARGDLGVEMPLEQIPLVQRRAIALCRQAAKPVIVATQML DSMVSDRRPTRAEVSDVANAVFDRADAVMLSAETSVGADPAHAVATMARIVVAAESGSGRGETPESALVE AVNGTDAVFGDAIAVAACEVGRRLGAAALCCFTRTGDTALRLARQRSPLPLLAFAHDDEVRGRLTFSWGV ESAVLAPTELAESMPGEVSRALLSMGACHPGDVVVVVSGSRSGVAGYTDSVRVLRVE
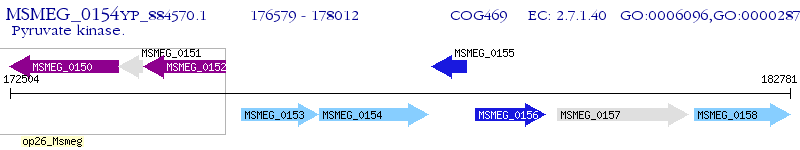
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0154 | pyk | - | 100% (477) | pyruvate kinase |
| M. smegmatis MC2 155 | MSMEG_3227 | pyk | e-149 | 56.54% (474) | pyruvate kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1643 | pykA | 1e-145 | 56.87% (473) | pyruvate kinase |
| M. gilvum PYR-GCK | Mflv_3591 | - | 1e-146 | 56.66% (473) | pyruvate kinase |
| M. tuberculosis H37Rv | Rv1617 | pykA | 1e-145 | 57.08% (473) | pyruvate kinase |
| M. leprae Br4923 | MLBr_01277 | pykA | 1e-148 | 57.93% (473) | pyruvate kinase |
| M. abscessus ATCC 19977 | MAB_2639c | - | 1e-146 | 58.14% (473) | pyruvate kinase |
| M. marinum M | MMAR_2420 | pykA | 1e-148 | 57.72% (473) | pyruvate kinase PykA |
| M. avium 104 | MAV_3170 | pyk | 1e-142 | 58.85% (452) | pyruvate kinase |
| M. thermoresistible (build 8) | TH_1097 | pykA | 1e-141 | 57.94% (447) | Probable pyruvate kinase pykA |
| M. ulcerans Agy99 | MUL_1598 | pykA | 1e-148 | 57.93% (473) | pyruvate kinase |
| M. vanbaalenii PYR-1 | Mvan_2824 | - | 1e-147 | 56.75% (474) | pyruvate kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1643|M.bovis_AF2122/97 -MTRRGKIVCTLGPATQRDDLVRALVEAGMDVARMNFSHGDYDDHKVAYE
Rv1617|M.tuberculosis_H37Rv -MTRRGKIVCTLGPATQRDDLVRALVEAGMDVARMNFSHGDYDDHKVAYE
MMAR_2420|M.marinum_M -MTRRGKIVCTLGPATQQDDLVKELVEAGMDVARMNFSHGDYPDHEAAYE
MUL_1598|M.ulcerans_Agy99 -MTRRGKIVCTLGPATQQDDLVKELVEAGMDVARMNFSHGDYADHEAAYE
MLBr_01277|M.leprae_Br4923 -MTRRGKIVCTLGPATQSDELIRALVEAGMDVARMNFSHGDYSDHKAVYD
MAV_3170|M.avium_104 ----------------------MALVEAGMDVARLNFSHGDYADHKAAYE
Mflv_3591|M.gilvum_PYR-GCK -MTRRGKIVCTLGPATASDEAVRQLVESGMDVARLNFSHGDYPDHEANYK
Mvan_2824|M.vanbaalenii_PYR-1 -MNRRGKIVCTLGPATATEEAVRQLVESGMDVARLNFSHGDYPDHEANYK
TH_1097|M.thermoresistible__bu -----------------------------MDVARLNFSHGTHADHERYYK
MAB_2639c|M.abscessus_ATCC_199 -MTRRGKIVCTLGPATGSAELVRALVEAGMDVARLNFSHGEHADHEQNYR
MSMEG_0154|M.smegmatis_MC2_155 MANRRAKIVCTLGPATATSSVLTELVDAGMDVARLNFSHSTHAEHSALYG
*****:****. : :*. *
Mb1643|M.bovis_AF2122/97 RVRVASDATGRAVGVLADLQGPKIRLGRFASGATHWAEGETVRITVGACE
Rv1617|M.tuberculosis_H37Rv RVRVASDATGRAVGVLADLQGPKIRLGRFASGATHWAEGETVRITVGACE
MMAR_2420|M.marinum_M RVRAASDATGRAVGVLADLQGPKIRLGRFADGPTYWADGDTVRITVEECE
MUL_1598|M.ulcerans_Agy99 RVRAASDATGRAVGVLADLQGPKIRLGRFADGPTYWADGDTVRITVEECE
MLBr_01277|M.leprae_Br4923 RVRVASDATGRAVGVLADLQGPKIRLGRFATGSTYWADGETVRITVADCH
MAV_3170|M.avium_104 RVRVASDATGRAVGVLADLQGPKIRLGRFATGPTYWADGETVRITVADCE
Mflv_3591|M.gilvum_PYR-GCK RVRAASDATGRAVGILADLQGPKIRLGRFADGPTVWRNGETIRITVDDFE
Mvan_2824|M.vanbaalenii_PYR-1 RVRAASDATGRAVGILADLQGPKIRLGRFADGPTMWANGETVRITVEDFV
TH_1097|M.thermoresistible__bu LVRAASDLTGRAVGILADLQGPKIRLGRFADGSTYWATGETVRITVDDIQ
MAB_2639c|M.abscessus_ATCC_199 WVRKASDESGRAVGVLADLQGPKIRLGRFATGSAHWATGEQVRITVEDVV
MSMEG_0154|M.smegmatis_MC2_155 MVREIAAQRGRVVGVLADLQGPKIRLGCFADGPVVWATGEHVTITTEDCP
** : **.**:************ ** *.. * *: : **.
Mb1643|M.bovis_AF2122/97 GSHDRVSTTYKRLAQDAVAGDRVLVDDGKVALVVDAVEGDDVVCTVVEGG
Rv1617|M.tuberculosis_H37Rv GSHDRVSTTYKRLAQDAVAGDRVLVDDGKVALVVDAVEGDDVVCTVVEGG
MMAR_2420|M.marinum_M GCRDRVSTTYKRLAQDAVAGDRVLVDDGKVGLIVDHIDGNDVVCTVVEGG
MUL_1598|M.ulcerans_Agy99 GCRDRVSTTYKRLAQDAVAGDRVLVDDGKVGLIVDHIDGNDVVCTVVEGG
MLBr_01277|M.leprae_Br4923 GSHDRVSTTYKNLARDAVVGDRVLVDDGKVGLVVSAIDGDDVVCTVTEGG
MAV_3170|M.avium_104 GSHDRVSTTYKQLAQDAAVGDRVLVDDGKVGLVVDGIEGDDVICTVVEGG
Mflv_3591|M.gilvum_PYR-GCK GTHDRVSTTYKRLAQDATPGDRVLVDDGNVGLVVEHIDGNDVVCTVTEGG
Mvan_2824|M.vanbaalenii_PYR-1 GNHDRVSTTYKRLAQDAKPGDRVLVDDGNVGLVVEHIDGNDVVCTVTEGG
TH_1097|M.thermoresistible__bu GTHDRVSTTYKNLAADAARGDRLLVDDGKVALVVTAVEGDDVVCEVVEGG
MAB_2639c|M.abscessus_ATCC_199 GTPDRVSTTYKQLAADAAVGDRLLVDDGKVGLTVEAIEGDDVVCRVTEGG
MSMEG_0154|M.smegmatis_MC2_155 GDHDRVSTTYAGLSQDVRAGDRLLVDDGRVDLRVVAVDGPDIRCEVVDGG
* ******* *: *. ***:*****.* * * ::* *: * *.:**
Mb1643|M.bovis_AF2122/97 PVSDNKGISLPGMNVTAPALSEKDIEDLTFALNLGVDMVALSFVRSPADV
Rv1617|M.tuberculosis_H37Rv PVSDNKGISLPGMNVTAPALSEKDIEDLTFALNLGVDMVALSFVRSPADV
MMAR_2420|M.marinum_M PVSNNKGISLPGMNVSAPALSEKDIDDLTFALNLGVDMVALSFVRSPSDV
MUL_1598|M.ulcerans_Agy99 PVSNNKGISLPGMNVSAPALSEKDIDDLTFALNLGVDMVALSFVRSPSDV
MLBr_01277|M.leprae_Br4923 PVSNNKGMSLPGMHVSAPALSDKDIEDLTFALNLGADLVALSFVRSPSDV
MAV_3170|M.avium_104 PVSNNKGISLPGMNVSAPALSDKDIEDLTFALELGVDLVALSFVRSPSDV
Mflv_3591|M.gilvum_PYR-GCK KVSNNKGMSLPGMNVSAPALSEKDIDDLEFALRLGVDLVALSFVRSPADV
Mvan_2824|M.vanbaalenii_PYR-1 KVSNNKGMSLPGMNVSAPALSEKDIDDLEFALRLGVDLVALSFVRSPADV
TH_1097|M.thermoresistible__bu PVSNNKGLSLPGMNVSVPALTEKDVEDLEFAIRLGVDMVALSFVRSPADV
MAB_2639c|M.abscessus_ATCC_199 PVSNNKGLSLPGMNVSVPALSEKDIADLEFALRLGVDLIALSFVRSPSDV
MSMEG_0154|M.smegmatis_MC2_155 PVSDHKGISLPNIPVSVPPLSDKDIEDLKFALELGADMIAMSFVRAPEEV
**::**:***.: *:.*.*::**: ** **:.**.*::*:****:* :*
Mb1643|M.bovis_AF2122/97 ELVHEVMDRIGRRVPVIAKLDKPEAIDNLEAIVLAFDAVMVARGDLGVEL
Rv1617|M.tuberculosis_H37Rv ELVHEVMDRIGRRVPVIAKLEKPEAIDNLEAIVLAFDAVMVARGDLGVEL
MMAR_2420|M.marinum_M ELVHEVMDRIGRRVPVIAKLEKPEAIDNLEAIVLAFDAVMVARGDLGVEL
MUL_1598|M.ulcerans_Agy99 ELVHEVMDRIGRRVPVIAKLEKPEAIDNLEAIVLAFDAVMVARGDLGVEL
MLBr_01277|M.leprae_Br4923 ELVHKAMDRVGRRVPVIAKLEKPEAVDNLEAIVLAFDAIMVARGDLGVEL
MAV_3170|M.avium_104 ELVHEVMDRVGRRVPVIAKLEKPEAVDNLEAIVLAFDAIMVARGDLGVEL
Mflv_3591|M.gilvum_PYR-GCK ELVHEIMDRVGRRVPVIAKLEKPEAIDNLEAIVLAFDAIMVARGDLGVEL
Mvan_2824|M.vanbaalenii_PYR-1 ELVHEVMDRVGRRVPVIAKLEKPEAIDNLEAIVLAFDAIMVARGDLGVEL
TH_1097|M.thermoresistible__bu ELVHEVMDRVGRRVPVIAKLEKPEAIDNLEAIVLAFDGVMVARGDLGVEL
MAB_2639c|M.abscessus_ATCC_199 ELVHAVMDRVGRRVPVIAKLEKPEAVANLEAIVLAFDAIMVARGDLGVEL
MSMEG_0154|M.smegmatis_MC2_155 ELAHKIMDEVGRRVPVIAKLEKPEAVSDLPAIVEAFDGLMVARGDLGVEM
**.* **.:**********:****: :* *** ***.:**********:
Mb1643|M.bovis_AF2122/97 PLEEVPLVQKRAIQMARENAKPVIVATQMLDSMIENSRPTRAEASDVANA
Rv1617|M.tuberculosis_H37Rv PLEEVPLVQKRAIQMARENAKPVIVATQMLDSMIENSRPTRAEASDVANA
MMAR_2420|M.marinum_M PLEEVPLVQKRAIQIARENAKPVIVATQMLDSMIENSRPTRAEASDVANA
MUL_1598|M.ulcerans_Agy99 PLEEVPLVQKRAIQIARENAKPVIVATQMLDSMIENSRPTRAEASDVANA
MLBr_01277|M.leprae_Br4923 PLEEVPLVQKRAIQMARENAKPVIVATQMLDSMIENSRPTRAEASDVANA
MAV_3170|M.avium_104 PLEEVPLVQKRAIQMARENAKPVIVATQMLDSMIENSRPTRAEASDVANA
Mflv_3591|M.gilvum_PYR-GCK PLEEVPLVQKRAIQMARENAKPVIVATQMLESMIENSRPTRAEASDVANA
Mvan_2824|M.vanbaalenii_PYR-1 PLEEVPLVQKRAIQMARENAKPVIVATQMLESMIENSRPTRAEASDVANA
TH_1097|M.thermoresistible__bu PLEEVPLVQKRAIQTARENAKPVIVATQMLESMIENSRPTRAEASDVANA
MAB_2639c|M.abscessus_ATCC_199 PLEDVPIVQKRAIQIARENARPVIVATQMLESMIENSRPTRAEASDVANA
MSMEG_0154|M.smegmatis_MC2_155 PLEQIPLVQRRAIALCRQAAKPVIVATQMLDSMVSDRRPTRAEVSDVANA
***::*:**:*** .*: *:*********:**:.: ******.******
Mb1643|M.bovis_AF2122/97 VLDGADALMLSGETSVGKYPLAAVRTMSRIICAVEENS-TAAPPLTHIPR
Rv1617|M.tuberculosis_H37Rv VLDGADALMLSGETSVGKYPLAAVRTMSRIICAVEENS-TAAPPLTHIPR
MMAR_2420|M.marinum_M VLDGADALMLSGETSVGKYPLAAVRTMSRIVCAVEENS-TAAPPLTHVPR
MUL_1598|M.ulcerans_Agy99 VLDGADALMLSGETSVGKYPLAAVRTMSRIVCAVEENS-TAAPPLTHVPR
MLBr_01277|M.leprae_Br4923 VLDGADALMLSEETSVGKYPLAAVKTMSRIVCAVEENS-TAAPPLTHVPR
MAV_3170|M.avium_104 VLDGADAVMLSGETSVGKYPLAAVRTMARIICAVEDNS-TAAPPLTHVPR
Mflv_3591|M.gilvum_PYR-GCK VLDGADAVMLSGETSVGKFPFETVRTMARIVSAVEDNS-VAVPALTHTPR
Mvan_2824|M.vanbaalenii_PYR-1 VLDGADAVMLSGETSVGKFPFETVRTMARIIKAVEENS-VAAPPLTHVPR
TH_1097|M.thermoresistible__bu VLDGTDAVMLSGETSVGKYPLEAVRTMASIVAAVEENS-TAAPPLTHLPR
MAB_2639c|M.abscessus_ATCC_199 VLDGADAVMLSGETSVGKYPLEAVRTMARIVSAVESHS-VSAPPLTHVPR
MSMEG_0154|M.smegmatis_MC2_155 VFDRADAVMLSAETSVGADPAHAVATMARIVVAAESGSGRGETPESALVE
*:* :**:*** ***** * :* **: *: *.*. * . .. : .
Mb1643|M.bovis_AF2122/97 TKRG-------VISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLP
Rv1617|M.tuberculosis_H37Rv TKRG-------VISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLP
MMAR_2420|M.marinum_M TKRG-------VISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLP
MUL_1598|M.ulcerans_Agy99 TKRG-------VISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLP
MLBr_01277|M.leprae_Br4923 TKRG-------VISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLP
MAV_3170|M.avium_104 TKRG-------VISYAARDIGERLDAKALVAFTQSGDTVKRLARLHTPLP
Mflv_3591|M.gilvum_PYR-GCK TKRG-------VISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLP
Mvan_2824|M.vanbaalenii_PYR-1 TKRG-------VISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLP
TH_1097|M.thermoresistible__bu TKRG-------VISYAARDIGERLDAKALVAFTQSGDTVRRLARLHTPLP
MAB_2639c|M.abscessus_ATCC_199 TKRG-------VISYAARDIGERLNAKALVAFTQSGDTVRRLARLHTPLP
MSMEG_0154|M.smegmatis_MC2_155 AVNGTDAVFGDAIAVAACEVGRRLGAAALCCFTRTGDTALRLARQRSPLP
: .* .*: ** ::*.**.* ** .**::***. **** ::***
Mb1643|M.bovis_AF2122/97 LLAFTAWPEVRSQLAMTWGTETFIVPKMQSTDGMIRQVDKSLLELARYKR
Rv1617|M.tuberculosis_H37Rv LLAFTAWPEVRSQLAMTWGTETFIVPKMQSTDGMIRQVDKSLLELARYKR
MMAR_2420|M.marinum_M LLAFTAWPEVRSQLAMTWGTETFIVPEMDSTDGMIRQVDKSLLELGRYKR
MUL_1598|M.ulcerans_Agy99 LLAFTAWPEVRSQLAMTWGTETFIVPEMDSTDGMIRQVDKSLLELGRYKR
MLBr_01277|M.leprae_Br4923 LLAFTAWPEVRSQLAMTWGTETFLVPKMLSTDGMIRQVDKSLLELGRYKR
MAV_3170|M.avium_104 LLAFTAWPEVRSQLAMTWGTETFIVPMMTSTDGMIRQVDKSLLELGRYRR
Mflv_3591|M.gilvum_PYR-GCK VLAFTSLPEVRSQLALTWGTETFIVPHIDTTDGMIRQVDQSMLNLGRYKR
Mvan_2824|M.vanbaalenii_PYR-1 VLAFTALPEVRSQLALTWGTETFIVPHIDTTDGMIRQVDQSMLNLGRYKR
TH_1097|M.thermoresistible__bu LLAFTSLPEIRSQLALTWGTETFIVPHIQTTDGMIRQVDQSLLQLGRYKR
MAB_2639c|M.abscessus_ATCC_199 VLAFTPLPEVRSQLALTWGTETFIVPVMETTDGMIQQVDTALLEMERYQR
MSMEG_0154|M.smegmatis_MC2_155 LLAFAHDDEVRGRLTFSWGVESAVLAPTELAESMPGEVSRALLSMGACHP
:***: *:*.:*:::**.*: ::. ::.* :*. ::*.: :
Mb1643|M.bovis_AF2122/97 GDLVVIVAGAPPGTVGSTNLIHVHRIGEDDV-
Rv1617|M.tuberculosis_H37Rv GDLVVIVAGAPPGTVGSTNLIHVHRIGEDDV-
MMAR_2420|M.marinum_M GDLVVIVAGAPPGTVGSTNLIHVHRIGEDDV-
MUL_1598|M.ulcerans_Agy99 GDLVVIVAGAPPGTVGSTNLIHVHRIGEDDV-
MLBr_01277|M.leprae_Br4923 GDLVVIVAGTPPGTVGSTNLIHVHRIGEDDV-
MAV_3170|M.avium_104 GDLVVIVAGAPPGTVGSTNLIHVHRIGEDDV-
Mflv_3591|M.gilvum_PYR-GCK GDLVVIVAGAPPGTVGSTNLIHVHRIGEDDV-
Mvan_2824|M.vanbaalenii_PYR-1 GDLVVIVAGAPPGTVGSTNLIHVHRIGEDDV-
TH_1097|M.thermoresistible__bu GDLVVIVAGAPPGTVGSTNMIHVHRIGEDDHF
MAB_2639c|M.abscessus_ATCC_199 GDLVVIVAGAPPGTVGSTNLIHVHRIGEDDH-
MSMEG_0154|M.smegmatis_MC2_155 GDVVVVVSGSRSGVAGYTDSVRVLRVE-----
**:**:*:*: .*..* *: ::* *: