For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSATVLDSIIEGVRADVAAREAVISLDEIKERAKAAPPPLNVMAALREPGIGVIAEVKRASPSRGALAS IGDPAELAQAYQDGGARVISVLTEQRRFNGSLDDLDAVRAAVSIPVLRKDFIVRPYQIHEARAHGADMLL LIVAALDQPVLESLLERTESLGMTALVEVHTEEEADRALKAGASVIGVNARDLKTLEVDRTVFSRIAPGL PSNVIRVAESGVRGTADLLAYAGAGADAVLVGEGLVTSGDPRSAVADLVTAGTHPSCPKPAR
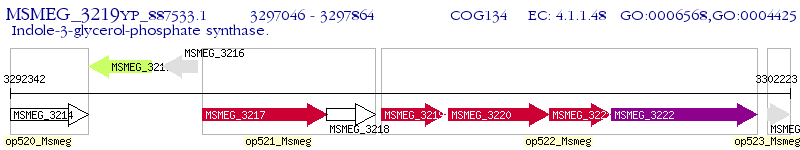
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3219 | trpC | - | 100% (272) | indole-3-glycerol-phosphate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1637 | trpC | 1e-130 | 87.13% (272) | indole-3-glycerol-phosphate synthase |
| M. gilvum PYR-GCK | Mflv_3600 | trpC | 1e-131 | 88.60% (272) | indole-3-glycerol-phosphate synthase |
| M. tuberculosis H37Rv | Rv1611 | trpC | 1e-130 | 87.13% (272) | indole-3-glycerol-phosphate synthase |
| M. leprae Br4923 | MLBr_01271 | trpC | 1e-127 | 85.66% (272) | indole-3-glycerol-phosphate synthase |
| M. abscessus ATCC 19977 | MAB_2645c | trpC | 1e-129 | 86.03% (272) | indole-3-glycerol-phosphate synthase |
| M. marinum M | MMAR_2413 | trpC | 1e-127 | 85.29% (272) | indole-3-glycerol phosphate synthase TrpC |
| M. avium 104 | MAV_3175 | trpC | 1e-129 | 86.76% (272) | indole-3-glycerol-phosphate synthase |
| M. thermoresistible (build 8) | TH_1076 | trpC | 1e-128 | 88.01% (267) | Probable indole-3-glycerol phosphate synthase trpC |
| M. ulcerans Agy99 | MUL_1591 | trpC | 1e-126 | 84.93% (272) | indole-3-glycerol-phosphate synthase |
| M. vanbaalenii PYR-1 | Mvan_2816 | trpC | 1e-136 | 91.91% (272) | indole-3-glycerol-phosphate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2413|M.marinum_M MSPATVLDSILEGVRADVAAREALVGMTEIKAAAAAAPPPLDVMAALRQP
MUL_1591|M.ulcerans_Agy99 MSPATVLDSILEGVRADVAAREALVGMTEIKAAAAAAPPPLDVMAALRQP
MAV_3175|M.avium_104 MSPATVLDSILEGVRADVAAREALISLSEIKAAAAAAPPPLDVMAALREP
Mb1637|M.bovis_AF2122/97 MSPATVLDSILEGVRADVAAREASVSLSEIKAAAAAAPPPLDVMAALREP
Rv1611|M.tuberculosis_H37Rv MSPATVLDSILEGVRADVAAREASVSLSEIKAAAAAAPPPLDVMAALREP
MLBr_01271|M.leprae_Br4923 MCPATVLDSILKGVRADVAAREACISLSEIKAAAAAAPAPLDAMAALREP
Mflv_3600|M.gilvum_PYR-GCK MGSATVLDSILEGVRADVAAREAVVSLAEVKARAERAPAPLDVMAALRAP
Mvan_2816|M.vanbaalenii_PYR-1 MGSATVLDSIIEGVRADVAAREAAVSMDEVKEQAKRAPAPLDVMAALRAS
MSMEG_3219|M.smegmatis_MC2_155 MSSATVLDSIIEGVRADVAAREAVISLDEIKERAKAAPPPLNVMAALREP
TH_1076|M.thermoresistible__bu -----VLDSIIEGVRADVAAREAEISLAEIKQRAKDAPPPLDVMAALREP
MAB_2645c|M.abscessus_ATCC_199 MSSGTVLDSILDGVRADVAAREAVVDLATVKAAAKAAPKPLDVMAALREP
*****:.*********** :.: :* * ** **:.***** .
MMAR_2413|M.marinum_M GIGVIAEVKRASPSAGSLAPITDPAKLARAYEDGGARIISVLTEGRRFQG
MUL_1591|M.ulcerans_Agy99 GIGVIAEVKRASPSAGSLAPITDPAKLARAYEDGGARIISVLTEGRRFQG
MAV_3175|M.avium_104 GIGVIAEVKRASPSAGSLATIADPAKLARAYEDGGARIISVLTEERRFHG
Mb1637|M.bovis_AF2122/97 GIGVIAEVKRASPSAGALATIADPAKLAQAYQDGGARIVSVVTEQRRFQG
Rv1611|M.tuberculosis_H37Rv GIGVIAEVKRASPSAGALATIADPAKLAQAYQDGGARIVSVVTEQRRFQG
MLBr_01271|M.leprae_Br4923 GIGVIAEVKRASPSVGSLATIADPAKLAQAYEDGGARIISVLTEERRFNG
Mflv_3600|M.gilvum_PYR-GCK GIAVIAEVKRASPSRGELASIADPAELASAYESGGARAISVLTEQRRFNG
Mvan_2816|M.vanbaalenii_PYR-1 GIAVIAEVKRASPSRGALASIADPAELARAYEDGGARIISVLTEQRRFNG
MSMEG_3219|M.smegmatis_MC2_155 GIGVIAEVKRASPSRGALASIGDPAELAQAYQDGGARVISVLTEQRRFNG
TH_1076|M.thermoresistible__bu GIAVIAEVKRASPSRGQLAAISDPAKLARAYEDGGARVISVLTEQRRFHG
MAB_2645c|M.abscessus_ATCC_199 GIAVIAEVKRASPSRGALAEISDPAELAKAYEDGGARIISVLTEGRRFNG
**.*********** * ** * ***:** **:.**** :**:** ***:*
MMAR_2413|M.marinum_M SLDDLDAVRAAVSIPLLRKDFVVQPYQIHEARAHGADMLLLIVAALDQSA
MUL_1591|M.ulcerans_Agy99 SLDDLDAVRAAVSIPLLRKDFVVQPYQIHEARAHGADMLLLIVAALDQSA
MAV_3175|M.avium_104 SLDDLDAVRAAVSIPVLRKDFVVQPYQIHEARAHGADMLLLIVAALEQSA
Mb1637|M.bovis_AF2122/97 SLDDLDAVRASVSIPVLRKDFVVQPYQIHEARAHGADMLLLIVAALEQSV
Rv1611|M.tuberculosis_H37Rv SLDDLDAVRASVSIPVLRKDFVVQPYQIHEARAHGADMLLLIVAALEQSV
MLBr_01271|M.leprae_Br4923 SLDDLDAVRAAVSVPVLRKDFVVQPYQIHEARAHGADMLLLIVAALDQSA
Mflv_3600|M.gilvum_PYR-GCK SLDDLDAVRAAVSIPVLRKDFIVRPYQIHEARAHGADLLLLIVAALEQPA
Mvan_2816|M.vanbaalenii_PYR-1 SLDDLDAVRAAVSIPVLRKDFIVRPYQIHEARAHGADMLLLIVAALEQPV
MSMEG_3219|M.smegmatis_MC2_155 SLDDLDAVRAAVSIPVLRKDFIVRPYQIHEARAHGADMLLLIVAALDQPV
TH_1076|M.thermoresistible__bu SLADLDAVRAAVSVPVLRKDFIIRPYQIHEARAHGADMLLLIVAALEQPA
MAB_2645c|M.abscessus_ATCC_199 SLDDLDSVRAAVSVPVLRKDFVISPYQIHEARAHGADLILLIVAALEQTA
** ***:***:**:*:*****:: *************::*******:*..
MMAR_2413|M.marinum_M LVSMLDRTESLGMTALVEVHTEQEADRALKAGAKVIGVNARDLTTLEVDR
MUL_1591|M.ulcerans_Agy99 LVSMLDRTESLGMTALVEVHTEQEADRALKAGAKVIGVNARDLTTLEVDR
MAV_3175|M.avium_104 LVSMLDRTESLGMTALVEVHTEEEADRALRAGAKVIGVNARDLATLEVDR
Mb1637|M.bovis_AF2122/97 LVSMLDRTESLGMTALVEVHTEQEADRALKAGAKVIGVNARDLMTLDVDR
Rv1611|M.tuberculosis_H37Rv LVSMLDRTESLGMTALVEVHTEQEADRALKAGAKVIGVNARDLMTLDVDR
MLBr_01271|M.leprae_Br4923 LMSMLDRTESLGMIALVEVRTEQEADRALKAGAKVIGVNARDLMTLEVDR
Mflv_3600|M.gilvum_PYR-GCK LESLLERTESLGMTALVEVHTEEEADRALQAGASLIGVNARNLKTLEVDR
Mvan_2816|M.vanbaalenii_PYR-1 LESLLERTESLGMTALVEVHTEEEADRALQAGASVIGVNARDLKTLEVDR
MSMEG_3219|M.smegmatis_MC2_155 LESLLERTESLGMTALVEVHTEEEADRALKAGASVIGVNARDLKTLEVDR
TH_1076|M.thermoresistible__bu LESMIERTHSLGMTPLVEVHTEEEADRALQAGAKVIGVNARDLKTLEVHR
MAB_2645c|M.abscessus_ATCC_199 LVSLLDRTESLGMTALVEVHTEEEADRALSAGASVIGVNARDLKTLEIDR
* *:::**.**** .****:**:****** ***.:******:* **::.*
MMAR_2413|M.marinum_M DCFARIAPGLPSNVIRIAESGVRGTNDLLAYAGAGADAVLVGEGLVKSGD
MUL_1591|M.ulcerans_Agy99 DCFARIASGLPSNVIRIAESGVRGTNDLLAYAGAGADAVLVGEGLVKSGD
MAV_3175|M.avium_104 DCFARIAPGLPSNVIRIAESGVRGTGDLLAYAGAGADAVLVGEGLVKSGD
Mb1637|M.bovis_AF2122/97 DCFARIAPGLPSSVIRIAESGVRGTADLLAYAGAGADAVLVGEGLVTSGD
Rv1611|M.tuberculosis_H37Rv DCFARIAPGLPSSVIRIAESGVRGTADLLAYAGAGADAVLVGEGLVTSGD
MLBr_01271|M.leprae_Br4923 DCFSRIAPGLPSNVIRIAESGVRGPADLLAYAGAGADAVLVGEGLVKSGD
Mflv_3600|M.gilvum_PYR-GCK DCFARIAPGLPTNVIKIAESGVRGTADLLAYAGAGADGVLVGEGLVTSGD
Mvan_2816|M.vanbaalenii_PYR-1 DCFARIAPGLPSNVIRVAESGVRGTADLLAYAGAGADAVLVGEGLVTSGD
MSMEG_3219|M.smegmatis_MC2_155 TVFSRIAPGLPSNVIRVAESGVRGTADLLAYAGAGADAVLVGEGLVTSGD
TH_1076|M.thermoresistible__bu DCFARIAPGLPSDVIRIAESGVRGPADLLAYAGAGADAVLVGEGLVTSGD
MAB_2645c|M.abscessus_ATCC_199 DCFSRIAPGLPSGVIRIAESGVRGTADLLAYAGAGADAVLVGEGLVTSGD
*:***.***:.**::*******. ***********.********.***
MMAR_2413|M.marinum_M PRAAVADLVTAGTHPSCPKPAR
MUL_1591|M.ulcerans_Agy99 PRAAVADLVTAGTHPSCPKPAR
MAV_3175|M.avium_104 PRAAVADLVTAGTHPSCPKPAR
Mb1637|M.bovis_AF2122/97 PRAAVADLVTAGTHPSCPKPAR
Rv1611|M.tuberculosis_H37Rv PRAAVADLVTAGTHPSCPKPAR
MLBr_01271|M.leprae_Br4923 PRAAVADLVTAGTHPSCPKPAR
Mflv_3600|M.gilvum_PYR-GCK PRSAVADLVTAGTHPSCPKPAR
Mvan_2816|M.vanbaalenii_PYR-1 PRSAVADLVTAGAHPSCPKPAR
MSMEG_3219|M.smegmatis_MC2_155 PRSAVADLVTAGTHPSCPKPAR
TH_1076|M.thermoresistible__bu PRSAVADLVTAGRHPSCPKPAR
MAB_2645c|M.abscessus_ATCC_199 PRGAVADLVTAGTHPSCPKPAR
**.********* *********