For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VATGATEAAGAGKRGLILLPAVDVVNGQAVRLVQGKAGSETDYGSAIEAAEAWQRDGAEWIHLVDLDAAF GRGSNRELLAEVVGKLDVAVELSGGIRDDESLRAALDTGCARVNIGTAALENPQWCARAIAEHGDRVAVG LDAQTDGAGGFRLRGRGWETDGGDLWEVLDRLDSEGCSRYVVTDVSKDGTLTGPNLDLLAAVAGRTDAPV IASGGVSSLDDLRAIATLTGVGVEGAIVGKALYAGRFTLPQALSAVSG
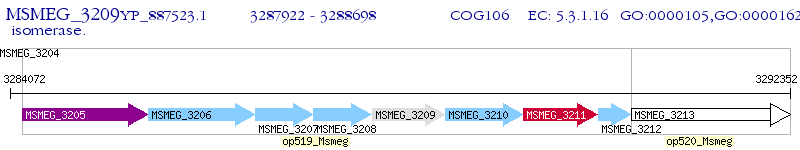
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3209 | - | - | 100% (258) | 1-(5-phosphoribosyl)-5- |
| M. smegmatis MC2 155 | MSMEG_3211 | hisF | 1e-17 | 24.50% (249) | imidazole glycerol phosphate synthase subunit HisF |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1629 | hisA | 1e-114 | 85.48% (241) | phosphoribosyl isomerase A |
| M. gilvum PYR-GCK | Mflv_3609 | - | 1e-113 | 83.82% (241) | 1-(5-phosphoribosyl)-5- |
| M. tuberculosis H37Rv | Rv1603 | hisA | 1e-114 | 85.48% (241) | phosphoribosyl isomerase A |
| M. leprae Br4923 | MLBr_01261 | hisA | 1e-108 | 79.25% (241) | phosphoribosyl isomerase A |
| M. abscessus ATCC 19977 | MAB_2666c | - | 1e-108 | 80.58% (242) | phosphoribosyl isomerase A |
| M. marinum M | MMAR_2399 | hisA | 1e-117 | 87.97% (241) | phosphoribosylformimino-5-aminoimidazole carboxamide |
| M. avium 104 | MAV_3183 | - | 1e-117 | 87.19% (242) | 1-(5-phosphoribosyl)-5- |
| M. thermoresistible (build 8) | TH_1159 | hisA | 1e-111 | 81.97% (244) | PROBABLE PHOSPHORIBOSYLFORMIMINO-5-AMINOIMIDAZOLE |
| M. ulcerans Agy99 | MUL_1576 | hisA | 1e-118 | 88.38% (241) | phosphoribosyl isomerase A |
| M. vanbaalenii PYR-1 | Mvan_2808 | - | 1e-113 | 84.65% (241) | 1-(5-phosphoribosyl)-5- |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2399|M.marinum_M --------MKEETVPLILLPAVDVVEGRAVRLVQGKAGSETEYGSALDAA
MUL_1576|M.ulcerans_Agy99 -------------MPLILLPAVDVVEGRAVRLVQGKAGSETEYGSALDAA
Mb1629|M.bovis_AF2122/97 ------------MMPLILLPAVDVVEGRAVRLVQGKAGSQTEYGSAVDAA
Rv1603|M.tuberculosis_H37Rv ------------MMPLILLPAVDVVEGRAVRLVQGKAGSQTEYGSAVDAA
MLBr_01261|M.leprae_Br4923 ------------MMSLILLPAVDVVEGRAVRLVQGKAGSENDYGSALDAA
MAV_3183|M.avium_104 --------------MLILLPAVDVVDGRAVRLVQGKAGSETEYGSALDAA
Mflv_3609|M.gilvum_PYR-GCK -----MNASS----KLILLPAVDVVEGRAVRLVQGQAGSETEYGSALDAA
Mvan_2808|M.vanbaalenii_PYR-1 --MTSVNPSSSKPSALILLPAVDVVEGRAVRLVQGQAGSETEYGSALDAA
TH_1159|M.thermoresistible__bu -------------VSLILLPAVDVVDGKAVRLVQGQAGSETEYGSALDAA
MSMEG_3209|M.smegmatis_MC2_155 MATGATEAAGAGKRGLILLPAVDVVNGQAVRLVQGKAGSETDYGSAIEAA
MAB_2666c|M.abscessus_ATCC_199 -------------MSLVLLPAVDVADGQAVRLVQGKAGSETTYGSPRDAA
*:*******.:*:*******:***:. ***. :**
MMAR_2399|M.marinum_M LGWQRDGAEWIHLVDLDAAFGRGSNRELLAEVVGKLDVAVELSGGIRDDD
MUL_1576|M.ulcerans_Agy99 LGWQRDGAEWIHLVDLDAAFGRGSNRELLAEVVGKLDVAVELSGGIRDDD
Mb1629|M.bovis_AF2122/97 LGWQRDGAEWIHLVDLDAAFGRGSNHELLAEVVGKLDVQVELSGGIRDDE
Rv1603|M.tuberculosis_H37Rv LGWQRDGAEWIHLVDLDAAFGRGSNHELLAEVVGKLDVQVELSGGIRDDE
MLBr_01261|M.leprae_Br4923 LCWQRDGADWIHLVDLDAAFGRGSNRELLSEMVGKLDVQVELSGGIRDDD
MAV_3183|M.avium_104 LGWQRDGAEWIHLVDLDAAFGRGSNRELLAEVVGKLDVQVELSGGIRDDD
Mflv_3609|M.gilvum_PYR-GCK MQWQSDGAEWIHLVDLDAAFGRGSNRELLADVVGRLDVAVELSGGIRDDE
Mvan_2808|M.vanbaalenii_PYR-1 MTWQRDGAEWIHLVDLDAAFGRGSNRELLAEVVGKLDVAVELSGGIRDDD
TH_1159|M.thermoresistible__bu LGWQRDGAEWIHLVDLDAAFGRGSNRELIAEVVGRLDVAVELSGGIRDHE
MSMEG_3209|M.smegmatis_MC2_155 EAWQRDGAEWIHLVDLDAAFGRGSNRELLAEVVGKLDVAVELSGGIRDDE
MAB_2666c|M.abscessus_ATCC_199 LAWQNDGAEWVHLVDLDAAFGRGSNRELLAQVVGELDVKVELSGGIRDDE
** ***:*:**************:**::::**.*** *********.:
MMAR_2399|M.marinum_M SLAAALATGCARVNLGTAALENPQWCARMIAEHGEKVAVGLDVQIV----
MUL_1576|M.ulcerans_Agy99 SLAAALATGCARVNLGTAALENPQWCARMIAEHGEKVAVGLDVQIV----
Mb1629|M.bovis_AF2122/97 SLAAALATGCARVNVGTAALENPQWCARVIGEHGDQVAVGLDVQII----
Rv1603|M.tuberculosis_H37Rv SLAAALATGCARVNVGTAALENPQWCARVIGEHGDQVAVGLDVQII----
MLBr_01261|M.leprae_Br4923 SLNAALATGCARVNLGTAACENPHWCAQVIAEHGDKIAVGLDVQIV----
MAV_3183|M.avium_104 SLAAALATGCARVNLGTAALENPQWCARAIGEHGDKVAVGLDVQTI----
Mflv_3609|M.gilvum_PYR-GCK SLAAALATGCARVNLGTAALENPQWCAKVVAEHGEKVAVGLDVKIE----
Mvan_2808|M.vanbaalenii_PYR-1 SLAAALATGCARVNLGTAALENPQWCAKVVAEHGDKVAVGLDVKIV----
TH_1159|M.thermoresistible__bu SLQAALATGCARVNLGTAALEDPQWCAEVIAEHGDKVAVGLDVKIDPSHP
MSMEG_3209|M.smegmatis_MC2_155 SLRAALDTGCARVNIGTAALENPQWCARAIAEHGDRVAVGLDAQTDG---
MAB_2666c|M.abscessus_ATCC_199 ALTAALATGCARVNLGTAALEDPEWCASAIARHGERVAVGLDVEIV----
:* *** *******:**** *:*.*** :..**:::*****.:
MMAR_2399|M.marinum_M DGKHRLRGRGWETDGGDLWEVLQRLDSEGCSRYVVTDVTKDGTLAGPNLE
MUL_1576|M.ulcerans_Agy99 GGKHRLRGRGWETDGGDLWEVLQRLDSEGCSRYVVTDVTKDGTLAGPNLE
Mb1629|M.bovis_AF2122/97 DGEHRLRGRGWETDGGDLWDVLERLDSEGCSRFVVTDITKDGTLGGPNLD
Rv1603|M.tuberculosis_H37Rv DGEHRLRGRGWETDGGDLWDVLERLDSEGCSRFVVTDITKDGTLGGPNLD
MLBr_01261|M.leprae_Br4923 DGQHRLRGRGWETDGGDLWDVLENLDRQGCSRFIVTDVTKDGTLDGPNLD
MAV_3183|M.avium_104 DGQHRLRGRGWETDGGDLWEVLERLERQGCSRYVVTDVTKDGTLGGPNLD
Mflv_3609|M.gilvum_PYR-GCK DGQYRLRGRGWETDGGDLWTVLERLDGEGCSRFVVTDVTKDGTLNGPNLD
Mvan_2808|M.vanbaalenii_PYR-1 DGQHRLRGRGWETDGGDLWTVLDRLDGEGCSRFVVTDVTKDGTLNGPNLE
TH_1159|M.thermoresistible__bu ESRHRLRGRGWESDGGDLWEVLERLNREGCSRYVVTDVTKDGTLKGPNLE
MSMEG_3209|M.smegmatis_MC2_155 AGGFRLRGRGWETDGGDLWEVLDRLDSEGCSRYVVTDVSKDGTLTGPNLD
MAB_2666c|M.abscessus_ATCC_199 EGAHRLRGRGWVSDGGDLWEVLERLRAQGCSRYVVTDVTKDGTLTGPNLE
. .******* :****** **:.* :****::***::***** ****:
MMAR_2399|M.marinum_M LLAGVAGRTEAPVIASGGVSSLDDLRAIATLTGQGVEGAIVGKALYAGRF
MUL_1576|M.ulcerans_Agy99 LLAGVAGRTEAPVIASGGVSSLDDLRAIATLTGQGVEGAIVGKALYAGRF
Mb1629|M.bovis_AF2122/97 LLAGVADRTDAPVIASGGVSSLDDLRAIATLTHRGVEGAIVGKALYARRF
Rv1603|M.tuberculosis_H37Rv LLAGVADRTDAPVIASGGVSSLDDLRAIATLTHRGVEGAIVGKALYARRF
MLBr_01261|M.leprae_Br4923 LLASVSDRTNVPVIASGGVSSLDDLRAIAKFTERGIEGAIVGKALYAERF
MAV_3183|M.avium_104 LLGAVADRTDAPVIASGGVSSLDDLRAIATLTGRGVEGAIVGKALYAGRF
Mflv_3609|M.gilvum_PYR-GCK LLAQVCERTDAPVIASGGVSSLDDLRAIATLTDRGVEGAIVGKALYAGRF
Mvan_2808|M.vanbaalenii_PYR-1 LLTQVCERTDAPVIASGGVSSLDDLRAIATLTDRGVEGAIVGKALYAGRF
TH_1159|M.thermoresistible__bu LLKAVCDRTDAPVIASGGVSSLDDLRAIAALTDRGVEGAIVGKALYAGRF
MSMEG_3209|M.smegmatis_MC2_155 LLAAVAGRTDAPVIASGGVSSLDDLRAIATLTGVGVEGAIVGKALYAGRF
MAB_2666c|M.abscessus_ATCC_199 LLAQVAGVANAPVIASGGVSSLDDLRAIAELTGAGVEGAIVGKALYAGRF
** *. ::.****************** :* *:*********** **
MMAR_2399|M.marinum_M TLPQALTAVRE
MUL_1576|M.ulcerans_Agy99 TLPQALTAVRE
Mb1629|M.bovis_AF2122/97 TLPQALAAVRD
Rv1603|M.tuberculosis_H37Rv TLPQALAAVRD
MLBr_01261|M.leprae_Br4923 TLPQALAVVRM
MAV_3183|M.avium_104 TLPQALAAVAE
Mflv_3609|M.gilvum_PYR-GCK TLPQALDAVRT
Mvan_2808|M.vanbaalenii_PYR-1 TLPQALDAVGP
TH_1159|M.thermoresistible__bu TLPEALAAVRQ
MSMEG_3209|M.smegmatis_MC2_155 TLPQALSAVSG
MAB_2666c|M.abscessus_ATCC_199 TLPDAIAAVS-
***:*: .*