For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPDRRGDRGRHQARKRAVDLLFEAEARGLTAEAVADSRAALAEDQDDVAPLNPYTVLVARGVTEHAAHID DLISAHLQGWTLERLPAVDRAILRVAVWELLHAEDVPEPVAVDEAVELAKELSTDESPGFVNGVLGQVML VTPQIRAASQAVRGTGPAEG
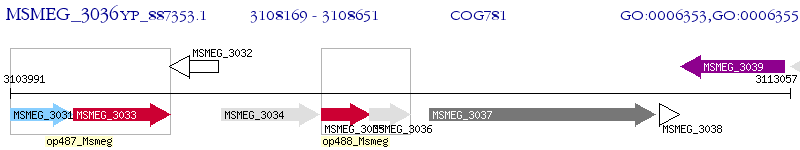
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3036 | nusB | - | 100% (160) | transcription antitermination protein NusB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2562c | nusB | 8e-68 | 80.52% (154) | transcription antitermination protein NusB |
| M. gilvum PYR-GCK | Mflv_3751 | nusB | 4e-72 | 84.62% (156) | transcription antitermination protein NusB |
| M. tuberculosis H37Rv | Rv2533c | nusB | 7e-67 | 79.87% (154) | transcription antitermination protein NusB |
| M. leprae Br4923 | MLBr_00523 | nusB | 1e-60 | 76.03% (146) | transcription antitermination protein NusB |
| M. abscessus ATCC 19977 | MAB_2836c | nusB | 2e-60 | 80.85% (141) | transcription antitermination protein NusB |
| M. marinum M | MMAR_2182 | nusB | 2e-64 | 78.95% (152) | N utilization substance protein NusB |
| M. avium 104 | MAV_3410 | nusB | 2e-65 | 82.31% (147) | transcription antitermination protein NusB |
| M. thermoresistible (build 8) | TH_0104 | nusB | 3e-73 | 87.66% (154) | N UTILIZATION SUBSTANCE PROTEIN NUSB (NUSB PROTEIN) |
| M. ulcerans Agy99 | MUL_1766 | nusB | 3e-64 | 78.95% (152) | transcription antitermination protein NusB |
| M. vanbaalenii PYR-1 | Mvan_2652 | nusB | 8e-72 | 85.81% (155) | transcription antitermination protein NusB |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3036|M.smegmatis_MC2_155 --------------MPDRRGD----RGRHQARKRAVDLLFEAEARGLTAE
TH_0104|M.thermoresistible__bu ----------VTDKKPERRAD----RGRHQARKRAVDLLFEAEARQITPA
MAB_2836c|M.abscessus_ATCC_199 MADIGSHSARAGDEPPGRRVSGSAKGGRRKARKRAVDLLFEAEAREMTAA
Mflv_3751|M.gilvum_PYR-GCK --------------MPDRKGD----RGRHQARKRAVDLLFEAEARGITAA
Mvan_2652|M.vanbaalenii_PYR-1 --------------MPDRKGD----RGRHQARKRAVDLLFEAEARGITAA
Mb2562c|M.bovis_AF2122/97 --------------MSD---R-KPVRGRHQARKRAVDLLFEAEVRGISAA
Rv2533c|M.tuberculosis_H37Rv --------------MSD---R-KPVRGRHQARKRAVALLFEAEVRGISAA
MMAR_2182|M.marinum_M --------------MPEGRPG-KPVKGRHQARKRAVDLMFEAEARGLGPG
MUL_1766|M.ulcerans_Agy99 --------------MPEGRPG-KPVKGRHQARKRAVDLMFEAEARGLGPG
MAV_3410|M.avium_104 --------------MS------KPLRGRHQARKRAVDLLFEAEARGLSPA
MLBr_00523|M.leprae_Br4923 --------------MS------NPVKGRHQARKRAVDLLFEAEARDLSPL
. **::****** *:****.* : .
MSMEG_3036|M.smegmatis_MC2_155 AVADSRAALAED-QDDVAPLNPYTVLVARGVTEHAAHIDDLISAHLQGWT
TH_0104|M.thermoresistible__bu EAAEQRNALAEK-QPDIAPLNPYTVKVVRGVTEHLAHIDDLISAHLQGWT
MAB_2836c|M.abscessus_ATCC_199 GVADERRALAAS-KPDIDPLNPYTVSVAHGVTEHAAHIDELISSHLQGWT
Mflv_3751|M.gilvum_PYR-GCK EVAEARNALADNGADDIAPLNPYTVTVARGVTDHAAHIDDLISAHLQGWT
Mvan_2652|M.vanbaalenii_PYR-1 EVAEARNALAEKRTDDIATLNPYTVTVAQGVTAHAAHIDDLISAHLQGWT
Mb2562c|M.bovis_AF2122/97 EVVDTRAALAEA-KPDIARLHPYTAAVARGVSEHAAHIDDLITAHLRGWT
Rv2533c|M.tuberculosis_H37Rv EVVDTRAALAEA-KPDIARLHPYTAAVARGVSEHAAHIDDLITAHLRGWT
MMAR_2182|M.marinum_M EIADLRTELAET-KPDVAPLHPYTAAVARGIDQHAAHIDDLITSHLQGWT
MUL_1766|M.ulcerans_Agy99 EIADLRTELAET-KPDVAPLHPYTAAVARGIGEHAAHIDDLITSHLQGWT
MAV_3410|M.avium_104 EVVDVRTGLADT-NPEVAPLQPYTAAVARGVGDHAAHIDDLISSHLQGWT
MLBr_00523|M.leprae_Br4923 EIIEVRSALAKS-KLDVAPLHPYTVVVAQGVSEHTARIDELIISHLQGWK
: * ** :: *:***. *.:*: * *:**:** :**:**.
MSMEG_3036|M.smegmatis_MC2_155 LERLPAVDRAILRVAVWELLHAEDVPEPVAVDEAVELAKELSTDESPGFV
TH_0104|M.thermoresistible__bu LERLPAVDRAILRVAVWELLHADDVPEPVAVDEAVELAKELSTDESPGFV
MAB_2836c|M.abscessus_ATCC_199 LARLPAVDRAILRVAVWELLHAQDVPEVVAVDEAVELAKQLSTDESPAFV
Mflv_3751|M.gilvum_PYR-GCK LDRLPAVDRAVLRVAVWELLHAVDVPEPVAVDEAVELAKQLSTDDSPGFV
Mvan_2652|M.vanbaalenii_PYR-1 LDRLPAVDRAILRVAVWELLHAEDVPEPVAVDEAVELAKQLSTDDSPGFV
Mb2562c|M.bovis_AF2122/97 LDRLPAVDRAILRVSVWELLHAADVPEPVVVDEAVQLAKELSTDDSPGFV
Rv2533c|M.tuberculosis_H37Rv LDRLPAVDRAILRVSVWELLHAADVPEPVVVDEAVQLAKELSTDDSPGFV
MMAR_2182|M.marinum_M LDRLPAVDRAILRVAVWELLYADDVPEPVAVDEAVELAKELSTDDSPSFV
MUL_1766|M.ulcerans_Agy99 LDRLPAVDRAILRVAVWELLYADDVPEPVAVDEAVELAKELSTDDSPSFV
MAV_3410|M.avium_104 LDRLPAVDRAILRVAVWELLYADDVPEPVAVDEAVQLAKELSTDDSPGFV
MLBr_00523|M.leprae_Br4923 LDRLPAVDRAILRVSIWELLYADDVPEPVAVDEAVELAKELSTDDSPGFV
* ********:***::****:* **** *.*****:***:****:**.**
MSMEG_3036|M.smegmatis_MC2_155 NGVLGQVMLVTPQIRAASQAVRGTGPAEG---------------------
TH_0104|M.thermoresistible__bu NGVLGQVMLVTPQIRAAAQAVRGTE-------------------------
MAB_2836c|M.abscessus_ATCC_199 NGVLGQLMLVTPHIRGSS-APSSTD-------------------------
Mflv_3751|M.gilvum_PYR-GCK NGVLGQVMLVTPQIRAAAAAVQASPDSGSATP------------------
Mvan_2652|M.vanbaalenii_PYR-1 NGVLGQVMLVTPQIRAAAAAMQGGHNGGSGT-------------------
Mb2562c|M.bovis_AF2122/97 NGVLGQVMLVTPQLRAAAQAVRGGA-------------------------
Rv2533c|M.tuberculosis_H37Rv NGVLGQVMLVTPQLRAAAQAVRGGA-------------------------
MMAR_2182|M.marinum_M NGVLGQVMLVTPQIRAAAAAVRQAV----PDESSRLPENQGSC-------
MUL_1766|M.ulcerans_Agy99 NGVLGQVMLVRPQIRAAAAAVRQAV----PDESSRLPENQGSC-------
MAV_3410|M.avium_104 NGVLGQVMLVTPQIRAAARAVRG------PRDT-----------------
MLBr_00523|M.leprae_Br4923 NGLLGKVMLVTPQIRAAAQAVQQAVRMAAGTSEDHVPQREPAAGQLGQDD
**:**::*** *::*.:: *
MSMEG_3036|M.smegmatis_MC2_155 -----------
TH_0104|M.thermoresistible__bu -----------
MAB_2836c|M.abscessus_ATCC_199 -----------
Mflv_3751|M.gilvum_PYR-GCK -----------
Mvan_2652|M.vanbaalenii_PYR-1 -----------
Mb2562c|M.bovis_AF2122/97 -----------
Rv2533c|M.tuberculosis_H37Rv -----------
MMAR_2182|M.marinum_M -----------
MUL_1766|M.ulcerans_Agy99 -----------
MAV_3410|M.avium_104 -----------
MLBr_00523|M.leprae_Br4923 SNGGQVAAVCR