For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGRFEDDAEVEDREVSPALTVGEGDIDASLRPRSLGEFIGQPRVREQLQLVLEGAKKRGGTPDHILLSGP PGLGKTSLAMIIAAELGSSLRVTSGPALERAGDLAAMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDF RVDVVVGKGPGATSIPLEVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEPVELERVLARSAGILGIE LGAEAGAEIARRSRGTPRIANRLLRRVRDFAEVRADGIITRDIAKSALEVYDVDELGLDRLDRAVLSALT RSFNGGPVGVSTLAVAVGEEATTVEEVCEPFLVRAGMIARTPRGRVATPLAWTHLGLQPPVTGIGQAGLF D
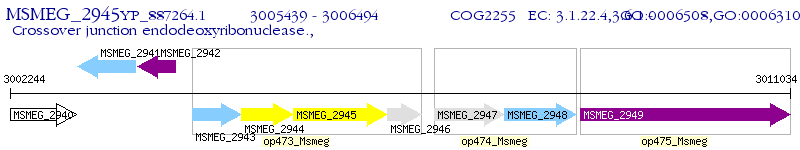
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2945 | ruvB | - | 100% (351) | Holliday junction DNA helicase B |
| M. smegmatis MC2 155 | MSMEG_3021 | - | 4e-10 | 28.83% (274) | recombination factor protein RarA |
| M. smegmatis MC2 155 | MSMEG_4671 | clpX | 4e-05 | 26.28% (156) | ATP-dependent protease ATP-binding subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2623c | ruvB | 1e-179 | 93.29% (343) | Holliday junction DNA helicase RuvB |
| M. gilvum PYR-GCK | Mflv_3824 | ruvB | 1e-179 | 91.09% (359) | Holliday junction DNA helicase B |
| M. tuberculosis H37Rv | Rv2592c | ruvB | 1e-179 | 93.29% (343) | Holliday junction DNA helicase RuvB |
| M. leprae Br4923 | MLBr_00483 | ruvB | 1e-174 | 90.64% (342) | Holliday junction DNA helicase RuvB |
| M. abscessus ATCC 19977 | MAB_2882c | ruvB | 1e-172 | 88.51% (348) | Holliday junction DNA helicase RuvB |
| M. marinum M | MMAR_2111 | ruvB | 1e-175 | 90.91% (341) | Holliday junction DNA helicase RuvB |
| M. avium 104 | MAV_3473 | ruvB | 1e-175 | 90.20% (347) | Holliday junction DNA helicase B |
| M. thermoresistible (build 8) | TH_1490 | ruvB | 1e-178 | 90.45% (356) | PROBABLE HOLLIDAY JUNCTION DNA HELICASE RUVB |
| M. ulcerans Agy99 | MUL_1716 | ruvB | 1e-175 | 90.62% (341) | Holliday junction DNA helicase RuvB |
| M. vanbaalenii PYR-1 | Mvan_2575 | ruvB | 0.0 | 92.68% (355) | Holliday junction DNA helicase B |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2945|M.smegmatis_MC2_155 ---MGRFE--DDAEVE--DREVSPALTVGEGDIDASLRPRSLGEFIGQPR
Mvan_2575|M.vanbaalenii_PYR-1 ---MGRFS--EDSADEPDEREMSAALTVGEGDIDASLRPRSLGEFIGQPR
Mflv_3824|M.gilvum_PYR-GCK ---MGRFSNADGPGDDADEREVTPALTVGEGDIDASLRPRSLGEFIGQPR
MMAR_2111|M.marinum_M ------------MSTDPDEREVSPALTVGDGDVDVSLRPRSLREFIGQPR
MUL_1716|M.ulcerans_Agy99 ------------MSTDPDEREVSPALTVGDGDVDVSLRPRSLREFIGQPR
MLBr_00483|M.leprae_Br4923 ------------MSEDYLDRDVSPALTVGEADIDVSLRPRSLREFIGQPR
Mb2623c|M.bovis_AF2122/97 ------------MTERS-DRDVSPALTVGEGDIDVSLRPRSLREFIGQPR
Rv2592c|M.tuberculosis_H37Rv ------------MTERS-DRDVSPALTVGEGDIDVSLRPRSLREFIGQPR
MAV_3473|M.avium_104 ---------MTAHDADWSDRDVSGALVPGEGDIDVSLRPRSLREFIGQPR
TH_1490|M.thermoresistible__bu ---MSRFD-AHAAEQGEPEREVSPALTVGEGDIDAGLRPRSLDEFIGQPR
MAB_2882c|M.abscessus_ATCC_199 MSRVNEFDDLDDGEDGFDNAHMSGALTVGENDVDAGLRPKSLREFIGQPR
: .:: **. *: *:*..***:** *******
MSMEG_2945|M.smegmatis_MC2_155 VREQLQLVLEGAKKRGGTPDHILLSGPPGLGKTSLAMIIAAELGSSLRVT
Mvan_2575|M.vanbaalenii_PYR-1 VREQLQLVLEGAKNRGGTPDHILLSGPPGLGKTSLAMIIAAELGSSLRVT
Mflv_3824|M.gilvum_PYR-GCK VREQLQLVLEGAKNRGGTPDHILLSGPPGLGKTSLAMIIAAELGSSLRVT
MMAR_2111|M.marinum_M VREQLQLVIQGAKNRGGTPDHILLSGPPGLGKTSLAMIIAAELGSSLRMT
MUL_1716|M.ulcerans_Agy99 VREQLQLVIQGAKNRGGTPDHILLSGPPGLGKTSLAVIIAAELGSSLRMT
MLBr_00483|M.leprae_Br4923 VREQLQLVIEGAKNRGATPDHILLSGPPGLGKTSLAMIIAAELGSSLRMT
Mb2623c|M.bovis_AF2122/97 VREQLQLVIEGAKNRGGTPDHILLSGPPGLGKTSLAMIIAAELGSSLRVT
Rv2592c|M.tuberculosis_H37Rv VREQLQLVIEGAKNRGGTPDHILLSGPPGLGKTSLAMIIAAELGSSLRVT
MAV_3473|M.avium_104 VREQLQLVIEGAKNRGGTPDHILLSGPPGLGKTSLAMIIAAELGSSLRVT
TH_1490|M.thermoresistible__bu VREQLQLVIEGAKNRGGTPDHILLSGPPGLGKTSLAMIIAAELGSSLRVT
MAB_2882c|M.abscessus_ATCC_199 VREQLELVLSGAKNRGGTPDHILLSGPPGLGKTSLAMIIAAELGSSLRVT
*****:**:.***:**.*******************:***********:*
MSMEG_2945|M.smegmatis_MC2_155 SGPALERAGDLAAMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDFRVD
Mvan_2575|M.vanbaalenii_PYR-1 SGPALERAGDLAAMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDFRVD
Mflv_3824|M.gilvum_PYR-GCK SGPALERAGDLAAMLSNLIEGDVLFIDEIHRIARPAEEMLYLAMEDFRVD
MMAR_2111|M.marinum_M SGPALERAGDLAAMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDFRVD
MUL_1716|M.ulcerans_Agy99 SGPALERAGDLAAMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDFRVD
MLBr_00483|M.leprae_Br4923 SGPALERAGDLAVMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDFRVD
Mb2623c|M.bovis_AF2122/97 SGPALERAGDLAAMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDFRVD
Rv2592c|M.tuberculosis_H37Rv SGPALERAGDLAAMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDFRVD
MAV_3473|M.avium_104 SGPALERAGDLAAMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDFRVD
TH_1490|M.thermoresistible__bu SGPALERAGDLAAMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDFRVD
MAB_2882c|M.abscessus_ATCC_199 SGPALERAGDLAAMLSNLVEHDVLFIDEIHRIARPAEEMLYLAMEDFRVD
************.*****:* *****************************
MSMEG_2945|M.smegmatis_MC2_155 VVVGKGPGATSIPLEVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEP
Mvan_2575|M.vanbaalenii_PYR-1 VVVGKGPGATSIPLEVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEP
Mflv_3824|M.gilvum_PYR-GCK VVVGKGPGATSIPLEVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEP
MMAR_2111|M.marinum_M VVVGKGPGATSIPLDVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEP
MUL_1716|M.ulcerans_Agy99 VVVGKGPGATSIPLDVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEP
MLBr_00483|M.leprae_Br4923 VIVGKGPGATSIPLEVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEP
Mb2623c|M.bovis_AF2122/97 VVVGKGPGATSIPLEVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEP
Rv2592c|M.tuberculosis_H37Rv VVVGKGPGATSIPLEVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEP
MAV_3473|M.avium_104 VVVGKGPGATSIPLEVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEP
TH_1490|M.thermoresistible__bu VVVGKGPGATSIPLEVAPFTLVGATTRSGALTGPLRDRFGFTAHMDFYEP
MAB_2882c|M.abscessus_ATCC_199 IVVGKGPGATSIPLEVAPFTLVGATTRSGSLTGPLRDRFGFTAHMDFYEP
::************:**************:********************
MSMEG_2945|M.smegmatis_MC2_155 VELERVLARSAGILGIELGAEAGAEIARRSRGTPRIANRLLRRVRDFAEV
Mvan_2575|M.vanbaalenii_PYR-1 SELERVLARSAGILGIELGAEAGAEIARRSRGTPRIANRLLRRVRDYAEV
Mflv_3824|M.gilvum_PYR-GCK AELERVLARSAGILGIELGAEAGAEIARRSRGTPRIANRLLRRVRDYAEV
MMAR_2111|M.marinum_M AELERVLVRSAGILGIQLGADAGAEIARRSRGTPRIANRLLRRVRDFAEV
MUL_1716|M.ulcerans_Agy99 AELERVLVRSAGILGIQLGADAGAEIARRSRGTPRIANRLLRRVRDFAEV
MLBr_00483|M.leprae_Br4923 TELEGVLARAAGILGIELGVEAGAEIARRSRGTPRIANRLLRRVRDFAEV
Mb2623c|M.bovis_AF2122/97 AELERVLARSAGILGIELGADAGAEIARRSRGTPRIANRLLRRVRDFAEV
Rv2592c|M.tuberculosis_H37Rv AELERVLARSAGILGIELGADAGAEIARRSRGTPRIANRLLRRVRDFAEV
MAV_3473|M.avium_104 AELQQVLARSAGILGIELGAEAAEEIARRSRGTPRIANRLLRRVRDFAEV
TH_1490|M.thermoresistible__bu AELEQVLARSAGILGIELGAEAAAEIARRSRGTPRIANRLLRRVRDYAEV
MAB_2882c|M.abscessus_ATCC_199 DELELVLARSAGILGIELGADAGTEIARRSRGTPRIANRLLRRVRDYAEV
**: **.*:******:**.:*. **********************:***
MSMEG_2945|M.smegmatis_MC2_155 RADGIITRDIAKSALEVYDVDELGLDRLDRAVLSALTRSFNGGPVGVSTL
Mvan_2575|M.vanbaalenii_PYR-1 RADGVITRDIAKYALEVYDVDELGLDRLDRAVLSALTRSFGGGPVGVSTL
Mflv_3824|M.gilvum_PYR-GCK RADGVITRDIAKYALEVYDVDELGLDRLDRAVLSALTRSFGGGPVGVSTL
MMAR_2111|M.marinum_M RADGVITRDVAKAALAVYDVDELGLDRLDRAVLSALTRSFSGGPVGVSTL
MUL_1716|M.ulcerans_Agy99 RADGVITRDVAKAALAVYDVDELGLDRLDRAVLSALTRSFSGGPVGVSTL
MLBr_00483|M.leprae_Br4923 RADGVITRDVAKAALAVYDVDELGLDRLDRAVLSALTRSFGGGPVGVSTL
Mb2623c|M.bovis_AF2122/97 RADGVITRDVAKAALEVYDVDELGLDRLDRAVLSALTRSFGGGPVGVSTL
Rv2592c|M.tuberculosis_H37Rv RADGVITRDVAKAALEVYDVDELGLDRLDRAVLSALTRSFGGGPVGVSTL
MAV_3473|M.avium_104 RADGVITRDVAKAALAVYDVDELGLDRLDRAVLTALTRSFGGGPVGVSTL
TH_1490|M.thermoresistible__bu RADGVITRDVAKAALEVYDVDELGLDRLDRAVLTALTRSFGGGPVGVSTL
MAB_2882c|M.abscessus_ATCC_199 RADGVITVDVAKSALAVYDVDELGLDRLDRAVLLALTRSFGGGPVGLSTL
****:** *:** ** ***************** ******.*****:***
MSMEG_2945|M.smegmatis_MC2_155 AVAVGEEATTVEEVCEPFLVRAGMIARTPRGRVATPLAWTHLGLQPP--V
Mvan_2575|M.vanbaalenii_PYR-1 AVAVGEEATTVEEVCEPFLVRAGMIARTPRGRVATAQAWTHLGMKPPANA
Mflv_3824|M.gilvum_PYR-GCK AVAVGEEATTVEEVCEPFLVRAGMIARTPRGRVATAQAWKHLGMTPPAGA
MMAR_2111|M.marinum_M AVAVGEEASTVEEVCEPFLVRAGMVARTPRGRVATALAWTHLGMTPPAGA
MUL_1716|M.ulcerans_Agy99 AVAVGEEASTVEEVCEPFLVRAGMVARTPRGRVATALAWTHLGMTPPAGA
MLBr_00483|M.leprae_Br4923 AVAVGEEATTVEEVCEPFLVRAGMVARTPRGRVATAQAWTYLCMTPPVGV
Mb2623c|M.bovis_AF2122/97 AVAVGEEAATVEEVCEPFLVRAGMVARTPRGRVATALAWTHLGMTPPVGA
Rv2592c|M.tuberculosis_H37Rv AVAVGEEAATVEEVCEPFLVRAGMVARTPRGRVATALAWTHLGMTPPVGA
MAV_3473|M.avium_104 AVAVGEEAATVEEVCEPFLVRAGMVARTPRGRVATAQAWTHLGMVPPAGA
TH_1490|M.thermoresistible__bu AVAVGEEATTVEEVCEPFLVRAGMIARTPRGRVATAQAWTHLGLSPPAGV
MAB_2882c|M.abscessus_ATCC_199 AVAVGEESTTVEEVCEPFLVRAGMLARTPRGRVATPLAWTHLGLTPPPQL
*******::***************:**********. **.:* : **
MSMEG_2945|M.smegmatis_MC2_155 TGIGQAGLFD--
Mvan_2575|M.vanbaalenii_PYR-1 VGLGQTGLFD--
Mflv_3824|M.gilvum_PYR-GCK GGLGQVGLFE--
MMAR_2111|M.marinum_M N---QPGLFE--
MUL_1716|M.ulcerans_Agy99 N---QPGLFE--
MLBr_00483|M.leprae_Br4923 TGLSQPGLFES-
Mb2623c|M.bovis_AF2122/97 S---QPGLFE--
Rv2592c|M.tuberculosis_H37Rv S---QPGLFE--
MAV_3473|M.avium_104 AGLGQPGLFD--
TH_1490|M.thermoresistible__bu TAFGQPALFEEP
MAB_2882c|M.abscessus_ATCC_199 LG--QTGLFD--
* .**: