For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRKLLAAALALTVTLAGCGLRSASGTVLDAGPGAIRHYDSLDGVPITVTAKDFTEQLILGNMVSIVLNAA GAEVTNMTNTPGSFGVRQAMLNGSATVSPEYTGNGWINYLGNEKPIPDPTAQWQAVNEADAVNNLTWLPP APMNNTYAFAIREAEGERLGVTKLSDLAGLPRNELTFCVESEFASRNDGFVPMLQTYGLSPDALGRVTHL DTGVIYTATADGDCNFGEVYTTDGRIPALDLRVLDDDRKFFPLYNLSVVVNSDLLRAHPELGEIFDELSP RLTNETMMALNARVDSDGDDPAIVARDWLVSEKLLS*
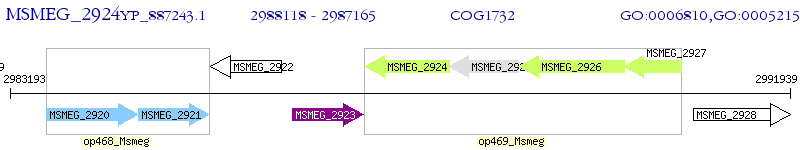
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2924 | - | - | 100% (317) | permease binding-protein component |
| M. smegmatis MC2 155 | MSMEG_6334 | - | 1e-12 | 28.77% (219) | ABC transporter, quaternary amine uptake transporter (QAT) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3785c | proX | 1e-15 | 26.88% (320) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3111 | - | 1e-78 | 47.65% (319) | substrate-binding region of ABC-type glycine betaine transport |
| M. tuberculosis H37Rv | Rv3759c | proX | 1e-15 | 26.88% (320) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1228 | - | 1e-136 | 77.74% (301) | putative ABC transporter |
| M. marinum M | MMAR_0032 | - | 1e-129 | 69.72% (317) | hypothetical protein MMAR_0032 |
| M. avium 104 | MAV_0317 | - | 3e-14 | 27.51% (269) | ABC transporter, quaternary amine uptake transporter (QAT) |
| M. thermoresistible (build 8) | TH_0699 | - | 1e-128 | 70.51% (312) | permease binding-protein component |
| M. ulcerans Agy99 | MUL_4377 | proX | 1e-17 | 28.40% (324) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. vanbaalenii PYR-1 | Mvan_5601 | - | 1e-18 | 28.21% (312) | substrate-binding region of ABC-type glycine betaine transport |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2924|M.smegmatis_MC2_155 --------------MTRKLLAAALALTVTLAGCGLRSASGTVLDAGPGAI
MAB_1228|M.abscessus_ATCC_1997 -------------------MVLACLVMMLQAGCGLQSASGAVLAAKPGLI
MMAR_0032|M.marinum_M MLAAVHRWSKCRRARRLAFAALAVLCVGLVASCGLRSATGAVLEAKPGLI
TH_0699|M.thermoresistible__bu -------------MVRGIALLCTVLLAAVTAGCGLQSSSGAVLKADPGSI
Mflv_3111|M.gilvum_PYR-GCK -------------MTRRAVLALLAVVTLTLSACGLGSGSAVPLRVGPGTI
Mb3785c|M.bovis_AF2122/97 -----------MRMLRRLRRATVAAAVWLATVCLVASCANADPLG--SAT
Rv3759c|M.tuberculosis_H37Rv -----------MRMLRRLRRATVAAAVWLATVCLVASCANADPLG--SAT
MUL_4377|M.ulcerans_Agy99 -----------MGTLRRMCRATLAAALCVVMVCSVAACGSSDPFG--TTS
MAV_0317|M.avium_104 ------------------------MAVLLTTMFVVAACSNSDPLG--AQV
Mvan_5601|M.vanbaalenii_PYR-1 -----------MICSRRTRRAAVAVAIALLAA-VLSACGSSNPLGGGEIS
: :
MSMEG_2924|M.smegmatis_MC2_155 RHYDSLDGVPITVTAKDFTEQLILGNMVSIVLNAAGAEVTNMTNTPGSFG
MAB_1228|M.abscessus_ATCC_1997 RHYDSLRDVNITVAAKDFTEQLVLGNIVAIILNAAGASVTNMTNTPGSFG
MMAR_0032|M.marinum_M RHYQSLEGVPITVAAKDFTEQLILGNMLSIVLHAAGARVTNLSNTPGSFG
TH_0699|M.thermoresistible__bu GYYESLDGVPITVAAKDFTEQLILGNMLATVLATAGAEVTNMSNTPGSFG
Mflv_3111|M.gilvum_PYR-GCK QPTPGLEGLKITVGSKEYTEQVIMGYILQFTLAAAGADVRDLTGIVGSRS
Mb3785c|M.bovis_AF2122/97 GSVK-----SIVVGSGDFPESQVIAEIYAQVLQANGFDVGRRLGIGSRET
Rv3759c|M.tuberculosis_H37Rv GSVK-----SIVVGSGDFPESQVIAEIYAQVLQANGFDVGRRLGIGSRET
MUL_4377|M.ulcerans_Agy99 CDIN-----SIVVGSGDFPESQIVAELYAQALRANGFEVGMRLGIGSRET
MAV_0317|M.avium_104 RSPD-----SIVVGSGDFPESQIIAEIYAQALQANGFDVGKRMGIGSRET
Mvan_5601|M.vanbaalenii_PYR-1 GDLK-----SIKVGSADFTESKIIAEIYAQALEANGFTISRQFGIGSRET
* * : ::.*. ::. : * : * : . .
MSMEG_2924|M.smegmatis_MC2_155 VRQAMLNGSATVSPEYTGNGWINYLGNEKPIPDPTAQWQAVNEAD-AVNN
MAB_1228|M.abscessus_ATCC_1997 VRQSMLKRVANVSPEYTGTGWINYLGHADPIKDPVAQWKAVNEED-AANR
MMAR_0032|M.marinum_M VRRALLTGEANIAPEYTGTGWINYLGHENPIKGAKQQWEAVNAAD-QVNG
TH_0699|M.thermoresistible__bu VREALLQGRADIAPEYTGTGWINYLGNDEPIKDEVEQWSAVKHAD-AVNG
Mflv_3111|M.gilvum_PYR-GCK TREAQLSGQVDIAYEFTGNAWINYLGHEKPIPDTREQYEAVRDEDLERND
Mb3785c|M.bovis_AF2122/97 YIPALKDHSIDLVPEYIGNLLLYFQPDATVTMLDAVELELYKRLP---GD
Rv3759c|M.tuberculosis_H37Rv YILALKDHSIDLVPEYIGNLLLYFQPDATVTMLDAVELELYKRLP---GD
MUL_4377|M.ulcerans_Agy99 YIPALKDHSIDLVPEYIGNLLLYFDPHATVTLLDAVELELYRRLP---GD
MAV_0317|M.avium_104 YIPAIKDHSIDLVPEYIGNLLLYFEPASSATTLDAVEQELHRRLP---AD
Mvan_5601|M.vanbaalenii_PYR-1 YIPAVRDHSIDLIPEYTGNLLQYFDPESAATTPDSVLLGLLKALP---GD
: : *: *. : .
MSMEG_2924|M.smegmatis_MC2_155 LTWLPPAPMNNTYAFAIREAEGERLGVTKLSDLAGLPRNE---LTFCVES
MAB_1228|M.abscessus_ATCC_1997 LVWLPPAPMNNTYAFAIRESEAERLAVTKLSDLRGLARND---LTFCVES
MMAR_0032|M.marinum_M LTWLPPAPMNNTYTLAIRESEAERLGVTKLSDLKRLPRSA---LTFCIES
TH_0699|M.thermoresistible__bu LTWLPPAPLNNTYAFAIRESEAERLGVTKMSDLHRLDRRD---LTFCVES
Mflv_3111|M.gilvum_PYR-GCK MVWLEPGPMDDTYALAASAQTIERTGVRTLSDYAALVMRDPAAARTCVDT
Mb3785c|M.bovis_AF2122/97 LSILTPSPASDTDTVTVTAATAARWNLKTIADLAPHSADV----KFAAPS
Rv3759c|M.tuberculosis_H37Rv LSILTPSPASDTDTVTVTAATAARWNLKTIADLAPHSADV----KFAAPS
MUL_4377|M.ulcerans_Agy99 LSVLTPSPASDTDTVTVTSATAGKWNLKTIADLAPHSANL----RFAAPS
MAV_0317|M.avium_104 LSILTPAPATDTDTVSVTSQAANSWNLRTIGDLAPHSADV----RFGAPS
Mvan_5601|M.vanbaalenii_PYR-1 LSILYPSPAEDKDTLAVSAETAQRWNLKSIADLAAHSAEV----KVGAPS
: * *.* :. :.: : .:.* :
MSMEG_2924|M.smegmatis_MC2_155 EFASRNDGFVPMLQTYGLSPDALGRVTHLDTG--VIYTATADG-DCNFGE
MAB_1228|M.abscessus_ATCC_1997 EFASRNDGFVPMLKTYGLSTGQLGRITNLDVG--VVYTATAKG-DCNFGE
MMAR_0032|M.marinum_M EFASRNDGFVPMLHAYGLDLEDLAKITTLDTG--VVYSATAQG-ECNFGE
TH_0699|M.thermoresistible__bu EFVNRNDGLVPMLAAYDLSRAALDRVTVLDTG--VIYTATANG-ECNFGE
Mflv_3111|M.gilvum_PYR-GCK EFRARQDGFPGMAAAYGFDP-ARAQTPILQVG--IIYQATADGTECDFGE
Mb3785c|M.bovis_AF2122/97 VFQTRPSGLPGLRHKYSLDIAPGNFVTINDGGGAVTVRALVEG-TATAAN
Rv3759c|M.tuberculosis_H37Rv AFQTRPSGLPGLRHKYSLDIAPGNFVTINDGGGAVTVRALVEG-TATAAN
MUL_4377|M.ulcerans_Agy99 AFQNRPSGLRGLREKYGLDISPQNFVTINDGGGAVTVRALLDG-TVDAAD
MAV_0317|M.avium_104 AFATRPAGLPGLRQKYGLDIKPGNFVAIDDGGGAVTVRALLEG-RVNAAN
Mvan_5601|M.vanbaalenii_PYR-1 EFQTRQTGLVGLKEKYGLDIAPANFVAISDGGGPATVKALTDG-TVTAAN
* * *: : *.:. . : * * .* .:
MSMEG_2924|M.smegmatis_MC2_155 VYTTDGRIPALDLRVLDDDRKFFPLYNLSVVVNSDLLRAHPELGEIFDEL
MAB_1228|M.abscessus_ATCC_1997 VFTTDGRIPALKLRVLQDDRHFFPLYNLTEVIDADLLAAHPELSEIFALL
MMAR_0032|M.marinum_M VFTTDGRIPALHLRLLEDDKHFFPLYNLTEVVDTSLINDHPELREIFAQL
TH_0699|M.thermoresistible__bu VFSTDGRIPALGLRVLDDDREFFPKYNMTEVVDTGLLEAHPEIAEIFAQL
Mflv_3111|M.gilvum_PYR-GCK VFTTDGRISALDLTVLTDDKQFFAHYNPSVTMKREFFEAHPEIAEVTAPV
Mb3785c|M.bovis_AF2122/97 LFSTSAAIPQNHLVVLEDPEHNFLAGNIVPLVNSRKKSDH--LKDVLDAV
Rv3759c|M.tuberculosis_H37Rv LFSTSAAIPQNHLVVLEDPEHNFLAGNIVPLVNSRKKSDH--LKDVLDAV
MUL_4377|M.ulcerans_Agy99 IFSTSPAIRQNHLVVLADPEHNFRAGNIVPLLNSQKKSDR--IKDVLDAV
MAV_0317|M.avium_104 IFTTSPAIPQHHLVALDDPEHNFIAGNVVPLVNSQKRSER--LKLVLDAV
Mvan_5601|M.vanbaalenii_PYR-1 IFSTSPAIERSALVVLEDPKNVFLAANVVPLVASQKMSNE--LKTVLDAV
:::*. * * * * .. * * : . : : :
MSMEG_2924|M.smegmatis_MC2_155 SPRLTNETMMALNARVD-SDGDDPAIVARDWLVSEKLLS----
MAB_1228|M.abscessus_ATCC_1997 NPRLTNDTMLALNARVD-SAGEDPAIVARDWLIDQGLLSR---
MMAR_0032|M.marinum_M NPRLTNDTMRELNAQVD-NNGRDPAIVAKDWLLQQKLLT----
TH_0699|M.thermoresistible__bu NPLITNEVMLALNAKVD-NDGADPALVARDWLIEQGLVR----
Mflv_3111|M.gilvum_PYR-GCK TAALTNEAITEMNKQVD-VDGRDPSVVARDWMVSQGFVTAE--
Mb3785c|M.bovis_AF2122/97 SAKLTTAGLAELNAAVSGNSGVDPDQAARKWVRDNGFDHPVRQ
Rv3759c|M.tuberculosis_H37Rv SAKLTTAGLAELNAAVSGNSGVDPDQAARKWVRDNGFDHPVRQ
MUL_4377|M.ulcerans_Agy99 SAKLTTEGVAELNASVSGNGGIDPNEAARKWLHDNGFDHPIRP
MAV_0317|M.avium_104 SAKLTTPGVAGLNAAVSGNSGVDPDQAARNWLRDNGLNHPIGP
Mvan_5601|M.vanbaalenii_PYR-1 SAKLTTEALIELNTSVEGNQGVDPDEAARKWISDNGFDTPIGK
.. :*. : :* *. * ** .*:.*: .: :