For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAPQSRGAYREAPGFGSIGEVSIDISLDPDTAFDTAPRPFHLAWCAQPPRRASMFAHITPNWFASVMGTG IVATAAVTLPVQASGLRAFATAVWILAAALLVALTAAFTTHWMRHRHHAVGYANHPVMAQFYGAPAMALL TVGAGSLLLGPAVIGESAAVRLGVALWVAGTGLGLAVAVVIPYRMITRCGREVAAVPAWMMPVVPPMVSA STGALLLGHLGPGQAGETLLMACYAMFGLSLLVGFVTVTMVYSRLMHGGLPEVQAIPTVWIVLGVIGQSI TATNLLAAHASSVVTDTSVVSALHAFGIVYGVVMGGFGAFVFCVAAALTVHAARRGLSFTLTWWSFTFPV GTCVTGASALGAATGAVVFSGLAVLLYVALLVAWVTVAMHTVRGVASGRLLAG
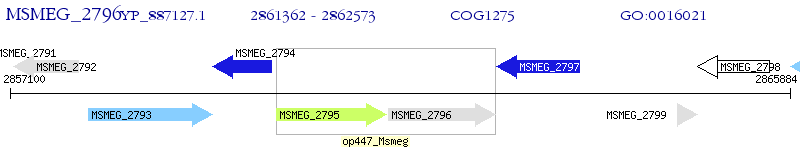
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2796 | - | - | 100% (403) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2282 | - | 4e-93 | 50.57% (350) | C4-dicarboxylate transporter/malic acid transport protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02529 | - | 3e-05 | 22.63% (327) | hypothetical protein MLBr_02529 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4962 | - | 2e-93 | 50.99% (355) | C4-dicarboxylate transporter/malic acid transport protein |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4035 | - | 3e-93 | 50.99% (355) | C4-dicarboxylate transporter/malic acid transport protein |
| M. vanbaalenii PYR-1 | Mvan_4384 | - | 7e-91 | 49.57% (351) | C4-dicarboxylate transporter/malic acid transport protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2282|M.gilvum_PYR-GCK -------------------------------MLRERACLHSDTPLTRG--
Mvan_4384|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4962|M.marinum_M --------------------------------------------------
MUL_4035|M.ulcerans_Agy99 MRTFRRSITAGSVFGSRSTLTHHGRGGNLPYAVRPAGGLAARTGRRRTGA
MSMEG_2796|M.smegmatis_MC2_155 -------------------------------MAPQSRGAYREAPGFGSIG
MLBr_02529|M.leprae_Br4923 -------------------------------MVYVFAEEFCEGPVTSGAV
Mflv_2282|M.gilvum_PYR-GCK ------------IFRALALAILSAMTVDANDDRPSILGYLGPN-------
Mvan_4384|M.vanbaalenii_PYR-1 ----------------------------MTNPPPTILGYLGPN-------
MMAR_4962|M.marinum_M ------------------MAVTNSPGMPSPHTRVEVLGNIGPN-------
MUL_4035|M.ulcerans_Agy99 QVAAPRGAEANSALGFSAMAVTNSPGTPSPHTRVEVLGNIGPN-------
MSMEG_2796|M.smegmatis_MC2_155 EVSIDISLDPDTAFDTAPRPFHLAWCAQPPR-RASMFAHITPN-------
MLBr_02529|M.leprae_Br4923 MPIVRVAILADSRLIEMALPTELPLREILPAVKRLVVPAASDNDSPLAAN
:. *
Mflv_2282|M.gilvum_PYR-GCK ----------------WFASVMGTGIVATAGASLPVHMPG------LRGF
Mvan_4384|M.vanbaalenii_PYR-1 ----------------WFASVMGTGIVATAGASLPVHVPG------LRGF
MMAR_4962|M.marinum_M ----------------WFASVMGTGIVAVAGATLPVHVVG------LRAF
MUL_4035|M.ulcerans_Agy99 ----------------WFASVMGTGIVAVAGATLPVHVVG------LRAF
MSMEG_2796|M.smegmatis_MC2_155 ----------------WFASVMGTGIVATAAVTLPVQASG------LRAF
MLBr_02529|M.leprae_Br4923 ASLHLSLAPIGGAPFSLDASLDTVGVVDGDLLALQPVPVGPAAPGIVEDI
**: .*:* :* * :. :
Mflv_2282|M.gilvum_PYR-GCK ATAVWVLSALWLVVLILAMLAHWLRNPTVARGHVRN---PTMAHFYGAAP
Mvan_4384|M.vanbaalenii_PYR-1 ALVVWVLSALWLLVLIFATFAHWLRNPVVARSHVRN---PQMAHFYGAAP
MMAR_4962|M.marinum_M TQVVWVIAAALLLALIILVGGHWLRHPTVARSHARN---PQMAHFYGAAP
MUL_4035|M.ulcerans_Agy99 TQVVWVIAAALLLALIVLVGGHWLRHPTVARSHARN---PQMAHFYGAAP
MSMEG_2796|M.smegmatis_MC2_155 ATAVWILAAALLVALTAAFTTHWMRHRHHAVGYANH---PVMAQFYGAPA
MLBr_02529|M.leprae_Br4923 ADAAMIFSTARLQSWGPTHIRRGALAATTAVTFAATGLGVTYRAVTGALT
: .. :::: * : * .. . ** .
Mflv_2282|M.gilvum_PYR-GCK MALLTVGSGALLVGEDLIGARLAVDLAWALWVAG-TLGGLFTAMTIPYLM
Mvan_4384|M.vanbaalenii_PYR-1 MALLTVGSGALLVGRDLIGERAAVDLAWVLWTTG-TLGALFTAMTIPYLM
MMAR_4962|M.marinum_M MALMTVGAGAVLAGGPLIGERLAVDLDWVLWTAG-TIGGLFTAVSIPFLM
MUL_4035|M.ulcerans_Agy99 MALMTVGADAVLAGGPLIGERLAVDLDWVLWTAG-TIGGLFTAVSIPFLM
MSMEG_2796|M.smegmatis_MC2_155 MALLTVGAGSLLLGPAVIGESAAVRLGVALWVAG-TGLGLAVAVVIPYRM
MLBr_02529|M.leprae_Br4923 GLLVVIVIAVLIALGGLVLRSRAARTGLVLSIAALVPIGAVFALAVPGIF
*:.: :: :: *. .* :. . . *: :* :
Mflv_2282|M.gilvum_PYR-GCK FTQYRVEPDAAFGGWLMPVVPP-------MVAAATGSLLIPHMAEGVGRA
Mvan_4384|M.vanbaalenii_PYR-1 FTQYRVEPDAAFGGWLMPVVPP-------MVAAATGSLLIPHMAEGPGRA
MMAR_4962|M.marinum_M FTQHRVEPDAAFGGWLMPVVPP-------MVSAATGALLLPHMPAGSGRE
MUL_4035|M.ulcerans_Agy99 FTQHRVEPDAAFGGWLMPVVPP-------MVSAATGALLLPHMPAGSGRE
MSMEG_2796|M.smegmatis_MC2_155 ITRCGREVAAVP-AWMMPVVPP-------MVSASTGALLLGHLGPGQAGE
MLBr_02529|M.leprae_Br4923 GPAQVLLASAGVTAWSLIALIVPGPERVRIVAFFTATVVIGVAVMLEAGA
. * .* : .: :*: *.:::: .
Mflv_2282|M.gilvum_PYR-GCK TMLYGCYAMFG--------LSLVASLIIITMVWSRLAHFGTSGTARVPTL
Mvan_4384|M.vanbaalenii_PYR-1 TMLYGCYAMFG--------LSLVASLIIITMVWSRLAHYGTSGTARVPTL
MMAR_4962|M.marinum_M TMLYGCYAMFG--------LSFVASLNIIAMIWSRLVLYGTSGTARVPTL
MUL_4035|M.ulcerans_Agy99 TMLYGCYAMFG--------LSFVASLNIIAMIWSRLVLYGTSGTARVPTL
MSMEG_2796|M.smegmatis_MC2_155 TLLMACYAMFG--------LSLLVGFVTVTMVYSRLMHGGLPEVQAIPTV
MLBr_02529|M.leprae_Br4923 ALLWQLTPLTIGCGLILAALLVTVEAAQLSALWARFPLPVIPAPGDPTPS
::* .: * . . :: :::*: . ..
Mflv_2282|M.gilvum_PYR-GCK WIVLGPLGQSITVAGLLGTDAAL-AVDENLADGMRVFAVLYG-VPVWGFA
Mvan_4384|M.vanbaalenii_PYR-1 WIVLGPLGQSITVAGLLGTDAAL-AVDARLADGMAVFAVLYG-VPVWGFA
MMAR_4962|M.marinum_M WIVLGPLGQSITAAGLLGAAAATGAVDHELAETMQAFAIIFG-VPVWGFA
MUL_4035|M.ulcerans_Agy99 WIVLGPLGQSITAAGLLGAAAATGAVDHELAETMQAFAIIFG-VPVWGFA
MSMEG_2796|M.smegmatis_MC2_155 WIVLGVIGQSITATNLLAAHASSVVTDTSVVSALHAFGIVYG-VVMGGFG
MLBr_02529|M.leprae_Br4923 APSFQVLEDLPRRVRISSAHQSGFIAAATLLSMLGSVAIALRPEAVSSVG
: : : . : .: : . : . : ..: : ...
Mflv_2282|M.gilvum_PYR-GCK VLWIGLATALTVRTLRRGMPFALTWWSLTFPVGTFVTGTTQLAVHTSLPA
Mvan_4384|M.vanbaalenii_PYR-1 VLWIALAAALTVRTLRRGMPFALTWWSLTFPVGTFVTGTAQLALHTGLPA
MMAR_4962|M.marinum_M MLWIALSTALTMRTLRRGMPFALTWWSLTFPVGTFVTGTSQLALHTHLPA
MUL_4035|M.ulcerans_Agy99 MLWIALSTALTVRTLRRGMPFALTWWSLTFPVGTFVTGTSQLALHTHLPA
MSMEG_2796|M.smegmatis_MC2_155 AFVFCVAAALTVHAARRGLSFTLTWWSFTFPVGTCVTGASALGAATGAVV
MLBr_02529|M.leprae_Br4923 WYLVAATAVAATLRARVWDSVACKAWLLAQPY-LVASVLLGLYTATGRYV
. ::. : * ..: . * :: * .: * * .
Mflv_2282|M.gilvum_PYR-GCK FRYAAAIAYIGLLCTWLLVAVRTLRVGIGRGLF-TPPGSDPVRAYKDPGP
Mvan_4384|M.vanbaalenii_PYR-1 FRYAAAVAYLGLLCTWLLVAVRTVRGGLRGGLF-APPAAGQIRAYKDPPT
MMAR_4962|M.marinum_M FRVAAAAAYAGLLATWVLVAIRTVRGSLRGNLLNLPPSSAPVKASKDPVL
MUL_4035|M.ulcerans_Agy99 FRVAAAAAYAGLLATWVLVAIRTVRGSLRGNLLNLPPSSAPFKASKDPVP
MSMEG_2796|M.smegmatis_MC2_155 FSGLAVLLYVALLVAWVTVAMHTVRGVASGRLLAG---------------
MLBr_02529|M.leprae_Br4923 AASAALLVLVVVVLAWAVVALNPRIASSDSYSLPLRRLLGMVASGLDASL
* :: :* **:.. :
Mflv_2282|M.gilvum_PYR-GCK -----------------
Mvan_4384|M.vanbaalenii_PYR-1 ATG--------------
MMAR_4962|M.marinum_M -----------------
MUL_4035|M.ulcerans_Agy99 -----------------
MSMEG_2796|M.smegmatis_MC2_155 -----------------
MLBr_02529|M.leprae_Br4923 IPAMAYLVSLFSWVLNR