For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLSAIAIVPAAPLLVPELMGAAAAEMAPLRDAALAAAAELPARWVSVGTGAADATFGPDQCGTFAGYGAA VPVTLGPVTNITPVELPLSALVAGWLRGRVAPQAVVDARVYAAATPAGDAVEIGRGLRTEIDATAEPVGV LVVADGAHTLSPSAPGGYRPESVAAQHALDEALASGDAAALASLPDVSVGRVAHEVLAGLAGAAPRTARE LYRGVPYGVGYFVGVWVP
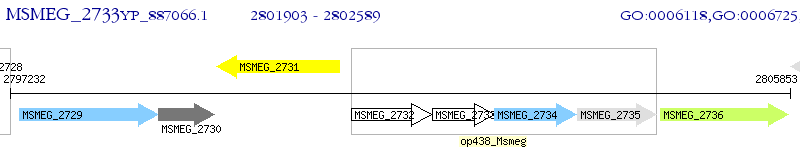
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2733 | - | - | 100% (228) | hypothetical protein MSMEG_2733 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2747c | - | 4e-67 | 56.71% (231) | hypothetical protein Mb2747c |
| M. gilvum PYR-GCK | Mflv_3963 | - | 5e-71 | 57.64% (229) | hypothetical protein Mflv_3963 |
| M. tuberculosis H37Rv | Rv2728c | - | 4e-67 | 56.71% (231) | hypothetical protein Rv2728c |
| M. leprae Br4923 | MLBr_00994 | - | 7e-64 | 56.90% (232) | hypothetical protein MLBr_00994 |
| M. abscessus ATCC 19977 | MAB_3045c | - | 3e-56 | 52.47% (223) | hypothetical protein MAB_3045c |
| M. marinum M | MMAR_1985 | - | 3e-70 | 58.87% (231) | hypothetical protein MMAR_1985 |
| M. avium 104 | MAV_3621 | - | 1e-71 | 61.30% (230) | hypothetical protein MAV_3621 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3371 | - | 1e-68 | 57.58% (231) | hypothetical protein MUL_3371 |
| M. vanbaalenii PYR-1 | Mvan_2433 | - | 5e-69 | 57.02% (228) | hypothetical protein Mvan_2433 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2747c|M.bovis_AF2122/97 ---MLSAIGIVPSAPVLVPELAGAAAAELADLGAAVIAAASLLPKSWIAV
Rv2728c|M.tuberculosis_H37Rv ---MLSAIGIVPSAPVLVPELAGAAAAELADLGAAVIAAASLLPKSWIAV
MLBr_00994|M.leprae_Br4923 ---MLSAIAIVPSAPVLVPELTGAAAAEVADLRSAVLAVAACLPPCWIVV
MMAR_1985|M.marinum_M ---MLSAIAIIPSAPVLVPELTGLAGAEVADLRSAVVAAASSLPNHWVGI
MUL_3371|M.ulcerans_Agy99 ---MLSAIAIIPSAPVLVPELTGLAGAEVADLRSAVVAAVSSLPNHWVGI
MAV_3621|M.avium_104 ---MLSVIAIVPSAPVLVPELAGTAADELAELSAATLAAAALLPDRWLIV
Mflv_3963|M.gilvum_PYR-GCK MVPVLSALAIVPSAPVMVPELASGAAAETADLRAAALEAVRGLPARWIAI
Mvan_2433|M.vanbaalenii_PYR-1 MELMLSAIVIVPSAPVMVPVLASSAAAETEDLRAAAVSAAAQLPRRWIAV
MSMEG_2733|M.smegmatis_MC2_155 ---MLSAIAIVPAAPLLVPELMGAAAAEMAPLRDAALAAAAELPARWVSV
MAB_3045c|M.abscessus_ATCC_199 ---MNLSIAICPSTPLLVPELGGEAAAETGELRAASVTVARSLPAQWIAI
: : * *::*::** * . *. * * * : .. ** *: :
Mb2747c|M.bovis_AF2122/97 GTGRADDVVRP-TDVGTFAGFGADVRVGLAP-QDGDGVAVPVELPLCALL
Rv2728c|M.tuberculosis_H37Rv GTGRADDVVRP-TDVGTFAGFGADVRVGLAP-QDGDGVAVPVELPLCALL
MLBr_00994|M.leprae_Br4923 GTGRADDVVGPGGCLGTFAGFGADVRVRLSP-QVGGEAELLVDFPVCALI
MMAR_1985|M.marinum_M GVGPEDQVAGP-DAVGTFAGFGVDVQVRLAP-ALGEQPGPPSDLPLCALL
MUL_3371|M.ulcerans_Agy99 GVDPEDQVAGP-EAVGTFAGFGVDVQVRLAP-ALGEQPGPPSDLPLCALL
MAV_3621|M.avium_104 GTGAADQELGD-DAVGTFAGFGMDVPVRLSPPADGRAETAPAALPLCVLL
Mflv_3963|M.gilvum_PYR-GCK GVAEQDGVYGP-DRAGTFGGYGVDIRVALSP---EATTSAPTALPLCALI
Mvan_2433|M.vanbaalenii_PYR-1 GVAAADGVHGP-ESVGTFAGYGVDVRVGLS----GPGSGEPTALPLCALI
MSMEG_2733|M.smegmatis_MC2_155 GTGAADATFGP-DQCGTFAGYGAAVPVTLGP----VTNITPVELPLSALV
MAB_3045c|M.abscessus_ATCC_199 GVG--DGQYHA-DMSGTFAGYGADVRVSLSD-----RMGKPAQLPLPVLI
*. * ***.*:* : * *. :*: .*:
Mb2747c|M.bovis_AF2122/97 TAWVRGQARPEARAQVHVYASDHGSDAAVARGRQLRADIDREPDPIGVLV
Rv2728c|M.tuberculosis_H37Rv TAWVRGQARPEARAQVHVYASDHGSDAAVARGRQLRADIDREPDPIGVLV
MLBr_00994|M.leprae_Br4923 AAWVRGQSQLDASAQVRVYCGDHDPDMALACGRQLRVEIEQAPDPIGVLV
MMAR_1985|M.marinum_M AGWVRGQVRPAATAKIQVYAHGRESEDALVQGKLLRAEIDRTADPVGVLV
MUL_3371|M.ulcerans_Agy99 AGWVRGQVRPAATAKIQVYAHGRESEDALVQGKLLRAEIDRTADPVGVLV
MAV_3621|M.avium_104 GGWLRGQVQPRAAARARIYRADYDADTAVRRGRQLRAELDALPEPVGVLV
Mflv_3963|M.gilvum_PYR-GCK CGWLRGQARPDARVEARAYRAEHAGPVALGFGARLREQIDADAADIGVVI
Mvan_2433|M.vanbaalenii_PYR-1 TAWLRGLARPQAVAEVWACRGDQEPGAAADLGRRLRAQLDAAPEEVGVLV
MSMEG_2733|M.smegmatis_MC2_155 AGWLRGRVAPQAVVDARVYAAATPAGDAVEIGRGLRTEIDATAEPVGVLV
MAB_3045c|M.abscessus_ATCC_199 AGWLRSRAEAD-EAEVRLVG---PATDALAFGRALRGEIERRPVPPGVLI
.*:*. . * * ** ::: . **::
Mb2747c|M.bovis_AF2122/97 VADGLNTLTPRAPGGYDPDGAGMQRALDDALASGDLAVLTRLPAQVLGRV
Rv2728c|M.tuberculosis_H37Rv VADGLNTLTPRAPGGYDPDGAGMQRALDDALASGDLAVLTRLPAQVLGRV
MLBr_00994|M.leprae_Br4923 VADGATTLTSSSPGGYDPSAADAELVLDDALASGDVAALTRLSCQISGRV
MMAR_1985|M.marinum_M VADGVNTLTPSAPGGYDPTGAPAQLLLDDALAGGDVAALARLPERVLGRP
MUL_3371|M.ulcerans_Agy99 VADGVNTLTPSAPGGYDPTGAPAQLLLDDALAGGDVAALARLPERVLGRP
MAV_3621|M.avium_104 VADGANTLTPAAPGGHHPGDAAAQQALDDALAGGDIAALRRLPGQILGRV
Mflv_3963|M.gilvum_PYR-GCK VADGANTLTPAAPGGYDEDSPTVQSALDDALAAGDCAALTRLPSGVLGRV
Mvan_2433|M.vanbaalenii_PYR-1 VADGANTLTPAAPGGHDPDSPRVQAALDDALATGDTAALMRLPSTVLGRV
MSMEG_2733|M.smegmatis_MC2_155 VADGAHTLSPSAPGGYRPESVAAQHALDEALASGDAAALASLPDVSVGRV
MAB_3045c|M.abscessus_ATCC_199 VADGANTLTDKAPGGYRPDAEAAQRAVVDALTRGDAASLRHLPDAITGRP
**** **: :***: : : :**: ** * * *. **
Mb2747c|M.bovis_AF2122/97 AFQVLAGLAEPGPRSAKEFYRGAPHGVGYFAGVWQP-
Rv2728c|M.tuberculosis_H37Rv AFQVLAGLAEPGPRSAKEFYRGAPHGVGYFAGVWQP-
MLBr_00994|M.leprae_Br4923 AFQVLAGLVEPGPRLAKELYRGAPYGVGYFVGVWQP-
MMAR_1985|M.marinum_M AFQVLAGLCGAGPRSATELYRGAPFGVGCFVGVWQP-
MUL_3371|M.ulcerans_Agy99 AFQVLAGLCGAGPRSATELYRGAPFGVGCFVGVWQP-
MAV_3621|M.avium_104 AFGVLAGLAEPGPRSAKELYRGAPYGVGYFAGAWQL-
Mflv_3963|M.gilvum_PYR-GCK AYQVLAGLAGTGPRTAAELYRGAPYGVGYFAGVWTP-
Mvan_2433|M.vanbaalenii_PYR-1 AYQVLAGLAEPGPRSAKELYRGAPYGVGYFVGVWTP-
MSMEG_2733|M.smegmatis_MC2_155 AHEVLAGLAGAAPRTARELYRGVPYGVGYFVGVWVP-
MAB_3045c|M.abscessus_ATCC_199 AYQVLAGLVDSDVVEARCLYRGSPYGVGYFAGIWRVS
*. ***** . * :*** *.*** *.* *