For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSVVRDTAAVMRVLDEDPVAGCMVAARVAEFGAEPGAIGGELWTRRRPSESLCYAGPNMIPLRGSLDDLK AFSDKAMSMARRCSSLVGRAELVLPMWERLQSLWGPARDVRAQQPLMALNSMPQCAIDPAVRPVRMEEID AYLVAAVDMFIGEVGVDPRIGDGGRGYRRRIASLIATGRAWARFERGEVVFKAEVGSQSPAVGQIQGVWV HPERRGHGLGTAGTAALAAAVVAGGRTASLYVNDYNTVARATYARIGFEQVGTFATVLLD
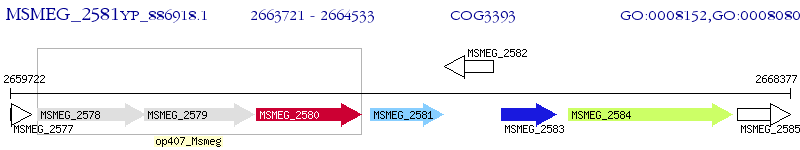
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2581 | - | - | 100% (270) | acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_2306 | - | 5e-05 | 31.91% (141) | putative acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_1026 | - | 5e-05 | 31.91% (141) | putative acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2892c | - | 1e-119 | 76.30% (270) | hypothetical protein Mb2892c |
| M. gilvum PYR-GCK | Mflv_4080 | - | 1e-127 | 80.37% (270) | GCN5-related N-acetyltransferase |
| M. tuberculosis H37Rv | Rv2867c | - | 1e-119 | 76.30% (270) | hypothetical protein Rv2867c |
| M. leprae Br4923 | MLBr_02193 | - | 7e-05 | 35.21% (71) | putative acetyltransferase |
| M. abscessus ATCC 19977 | MAB_3168c | - | 7e-92 | 62.12% (264) | hypothetical protein MAB_3168c |
| M. marinum M | MMAR_1839 | - | 1e-122 | 77.78% (270) | hypothetical protein MMAR_1839 |
| M. avium 104 | MAV_3724 | - | 1e-120 | 77.41% (270) | acetyltransferase |
| M. thermoresistible (build 8) | TH_1398 | - | 1e-126 | 80.37% (270) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2088 | - | 1e-122 | 77.41% (270) | hypothetical protein MUL_2088 |
| M. vanbaalenii PYR-1 | Mvan_2263 | - | 1e-124 | 79.03% (267) | GCN5-related N-acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2892c|M.bovis_AF2122/97 --MSAPPISRLVGERQVSVVRDAA---AVWRVLDDDPIESCMVAAR----
Rv2867c|M.tuberculosis_H37Rv --MSAPPISRLVGERQVSVVRDAA---AVWRVLDDDPIESCMVAAR----
MMAR_1839|M.marinum_M --MSAPPIMRLVGERRVSVVRDVA---ALWRVLDENPVESCMVAAR----
MUL_2088|M.ulcerans_Agy99 --MSAPPIMRLVGERRVSVVRDVA---ALWRVLDENPVESCMVAAR----
MAV_3724|M.avium_104 ----------------MSVVRDAA---AVWRVLDDDPVASCMVAAR----
Mflv_4080|M.gilvum_PYR-GCK --MSAPPLFRLVDERRVSVVRDIQ---AVRQVLDDDPVASCMVASR----
Mvan_2263|M.vanbaalenii_PYR-1 --MSAPPLFRLVDERRVSVVRDIRDMVAVRQVLDDDPVAACMVASR----
TH_1398|M.thermoresistible__bu VAMSAPPVFRLADERRVSLVRDPV---AVRAVLDEDPVASCMVAAR----
MSMEG_2581|M.smegmatis_MC2_155 ----------------MSVVRDTA---AVMRVLDEDPVAGCMVAAR----
MAB_3168c|M.abscessus_ATCC_199 ----------------MLGLSDTE---AVRAVLDIDPVASCMLAAR----
MLBr_02193|M.leprae_Br4923 ---MVLNWRFALSADEQRLVREIIS--AATEFDEVSPVGEQVLRELGYDR
: : * . : .*: ::
Mb2892c|M.bovis_AF2122/97 ----VADHGIDPNAIGGELWTRRGAHESLCFAGANLIPLRGGPIDLNAFA
Rv2867c|M.tuberculosis_H37Rv ----VADHGIDPNAIGGELWTRRGAHESLCFAGANLIPLRGGPIDLNAFA
MMAR_1839|M.marinum_M ----VADYGIEPNSLGGELWTRDGVDESLCFAGANLIPLRGELIDLNAFA
MUL_2088|M.ulcerans_Agy99 ----VADYGIEPNSLGGDLWTRDGVDESLCFAGANLIPLRGELIDLNAFA
MAV_3724|M.avium_104 ----VADHGIDPNAIGGELWTLRGADESLCFAGANLVPLRGAPGDLTAFA
Mflv_4080|M.gilvum_PYR-GCK ----VAEHGIDPAAIGGELWTRRRANESLCFAGPNLIPLRGGAGDLNAFA
Mvan_2263|M.vanbaalenii_PYR-1 ----VAEHGIDPSAIGGELWTRRRATESLCFAGANLIPLRGGAGDLNAFA
TH_1398|M.thermoresistible__bu ----VAEHGVDPAAIGGELWTRRRVDEALCFAGANLIPLRGRLDDLHAFA
MSMEG_2581|M.smegmatis_MC2_155 ----VAEFGAEPGAIGGELWTRRRPSESLCYAGPNMIPLRGSLDDLKAFS
MAB_3168c|M.abscessus_ATCC_199 ----VEARGADPTAIGGEIWSTGVLADSLCFVGTTIIPLAGGPEATRLFA
MLBr_02193|M.leprae_Br4923 TEHLLVTDSRPYAPIIGYLNLSSPRDAGVAMAELVVHPRERRRGVGAAMV
: . .: * : .:. . : * :
Mb2892c|M.bovis_AF2122/97 DVAMSTPRRCSSLVGRADLVLPMWQRLEPVWGPARDVRDNQPLMALATHP
Rv2867c|M.tuberculosis_H37Rv DVAMSTPRRCSSLVGRADLVLPMWQRLEPVWGPARDVRDNQPLMALATHP
MMAR_1839|M.marinum_M DEAKSTARRCSSLVGRADLVLPMWQRLESAWGPARDVRDHQPLMAINTHP
MUL_2088|M.ulcerans_Agy99 DEAKSTARRCSSLVGRADLVLPMWQRLESAWGPARDVRDHQPLMAINTHP
MAV_3724|M.avium_104 DEAMRGARRCSSLVGRADLVMPMWERLEAAWGPARDVRDRQPLLALSSYP
Mflv_4080|M.gilvum_PYR-GCK DKAMSSVRRCSSLVGRAELVLPMWQRLEPVWGPARDVRAHQPLMALNTAP
Mvan_2263|M.vanbaalenii_PYR-1 DKAMSSARRCSSLVGRAELVLPMWQRLEPVWGPARDVRAHQPLMALSTTP
TH_1398|M.thermoresistible__bu DKAVGQPRRCSSLVGRAELVLPMWQRLESGWGPARDVRDEQPLMALSTEP
MSMEG_2581|M.smegmatis_MC2_155 DKAMSMARRCSSLVGRAELVLPMWERLQSLWGPARDVRAQQPLMALNSMP
MAB_3168c|M.abscessus_ATCC_199 ERAIEQHRICPSLVGPADVVLDMWCHLDPVWGPAREVRACQPLLAMLDAP
MLBr_02193|M.leprae_Br4923 RAALAKTGGRNRFWAHGTLASARATASVLGLVPVRELVQMQRSLRTIPDP
* : . . :. *.*:: * : *
Mb2892c|M.bovis_AF2122/97 SCAIDTGVRQ---VRPEELDSYLVAAVDMFIGEVG----VDPRLGDGGRG
Rv2867c|M.tuberculosis_H37Rv SCAIDTGVRQ---VRPEELDSYLVAAVDMFIGEVG----VDPRLGDGGRG
MMAR_1839|M.marinum_M SCAIDPEVRQ---VRPEELDAYLVAAVDMFIGEVG----IDPRLGDGGRG
MUL_2088|M.ulcerans_Agy99 SCAIDPEVRQ---VRPEELDAYLVAAVDMFIGEVG----IDPRLGDGGRG
MAV_3724|M.avium_104 SCDIDTAVRQ---VRPEELDAYLVAAVDMFIGEVG----VDPRIGDGGRG
Mflv_4080|M.gilvum_PYR-GCK SCPTDPAVRP---VRMEELDAYLVAAIDMFIGEVG----IDPRLGDGGRG
Mvan_2263|M.vanbaalenii_PYR-1 LSAIDPAVRQ---VRIEELDAYLVAAIDMFIGEVG----IDPRIGDGGRG
TH_1398|M.thermoresistible__bu CCPIDPAVRP---VRMEELDAYLVAAIDMFIGEVG----IDPRLGDGGRG
MSMEG_2581|M.smegmatis_MC2_155 QCAIDPAVRP---VRMEEIDAYLVAAVDMFIGEVG----VDPRIGDGGRG
MAB_3168c|M.abscessus_ATCC_199 TCVVDPAVRQ---VRAEELDEYLNASVQMFIGEMG----VDPRNGDGGRG
MLBr_02193|M.leprae_Br4923 MVPDQLGVWVRTYVGTVDDAELLRVNNAAFAGHPEQGGWTATQLAERRSE
: * * : * . * *. .: .:
Mb2892c|M.bovis_AF2122/97 YRRRVAGLIAAGRAWARFEHGQVI--FKAEVGSQSPAVGQIQGVWVHPEW
Rv2867c|M.tuberculosis_H37Rv YRRRVAGLIAAGRAWARFEHGQVI--FKAEVGSQSPAVGQIQGVWVHPEW
MMAR_1839|M.marinum_M YRRRVASLIAAGRAWARFEHGEVI--FKAEVGSQSPAVGQIQGVWVHPER
MUL_2088|M.ulcerans_Agy99 YRRRVASLIAAGRAWARFEHGEVI--FKAEVGSQSPAVGQIQGVWVHPER
MAV_3724|M.avium_104 YRRRVASLITAGRAWARFEHGQVV--FKAEVGSQSPGVGQIQGVWVHPEW
Mflv_4080|M.gilvum_PYR-GCK YRRRVAGLIAAGRAWARFERGEVV--YKAEVGSQSPAVGQIQGVWVHPER
Mvan_2263|M.vanbaalenii_PYR-1 YRRRVAGLIAAGRAWARFDRGEVV--FKAEVGSQSPAVGQIQGVWVHPDQ
TH_1398|M.thermoresistible__bu YRRRVAGLIAAGRAWARFERGEVV--FKAEVGSQSPAVGQIQGVWVHPDW
MSMEG_2581|M.smegmatis_MC2_155 YRRRIASLIATGRAWARFERGEVV--FKAEVGSQSPAVGQIQGVWVHPER
MAB_3168c|M.abscessus_ATCC_199 YRRRVAGLIEAGRAWARFEDGRVI--FKAEIGSQSRSVSQIQGVWVDPEF
MLBr_02193|M.leprae_Br4923 PWFDPAGLFLAFGDSSSNQPGKLLGFHWTKVHAAHPGLGEVYVLGVDPSA
*.*: : : : *.:: . ::: : .:.:: : *.*.
Mb2892c|M.bovis_AF2122/97 RGIGLG----TAGTATLAAVIVG--SGRIASLYVNSFNTVARAAYARVGF
Rv2867c|M.tuberculosis_H37Rv RGIGLG----TAGTATLAAVIVG--SGRIASLYVNSFNTVARAAYARVGF
MMAR_1839|M.marinum_M RGLGLG----TAGTATLAAVTVG--SGRIASLYVNNFNTVARAAYTRVGF
MUL_2088|M.ulcerans_Agy99 RGLGLG----TAGTATLAAVTVG--SGRIASLYVNNFNTVARAAYTRVGF
MAV_3724|M.avium_104 RGLGLG----TAGTATVAAVIVG--SGRTASLYVNDFNTVARAAYARVGF
Mflv_4080|M.gilvum_PYR-GCK RGCGLG----TAGTASVAAAVVG--AGRIASLYVNSFNTVARATYARIGF
Mvan_2263|M.vanbaalenii_PYR-1 RGRGIG----TAGTAAVAAAIVA--GGRIASLYVNSFNTVARATYARIGF
TH_1398|M.thermoresistible__bu RGRGLG----SAGTATLASAVVR--GGRIASLYVNSFNTVARATYARVGF
MSMEG_2581|M.smegmatis_MC2_155 RGHGLG----TAGTAALAAAVVA--GGRTASLYVNDYNTVARATYARIGF
MAB_3168c|M.abscessus_ATCC_199 RGRGLG----TAGVAAIAAAVHR--SGRIPSLYVNSFNEVARASYAKAGF
MLBr_02193|M.leprae_Br4923 QGRGLGQMLTSIGIASLAQRLVGPSAEPTVMLYVESDNVAAARTYERLGF
:* *:* : * *::* . ***:. * .* :* : **
Mb2892c|M.bovis_AF2122/97 KEIGTFATVLLD----
Rv2867c|M.tuberculosis_H37Rv KEIGTFATVLLD----
MMAR_1839|M.marinum_M KEVGTFATVLLD----
MUL_2088|M.ulcerans_Agy99 KEVGTFATVLLD----
MAV_3724|M.avium_104 AEIGTFATVLLD----
Mflv_4080|M.gilvum_PYR-GCK TEIASFATVLLD----
Mvan_2263|M.vanbaalenii_PYR-1 TEIASFATVLLD----
TH_1398|M.thermoresistible__bu QTVGTFATVLLD----
MSMEG_2581|M.smegmatis_MC2_155 EQVGTFATVLLD----
MAB_3168c|M.abscessus_ATCC_199 TRIGTFATVLVS----
MLBr_02193|M.leprae_Br4923 TTYSVDTAYALARIDD
. :: :