For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRRRTVSTYSGEKMTQSSTKVFGWQRTGTRPLLITLIDGPNMPNLGNRNKRVYGPISSITALQDHVRELG EGMGVTIEALASNHEGEILDAIHASAARSDAYILNPAGLTKTGEPTRHALEETGRPWVEVHFANISAPPW SERGLPGGPWESNFTRSATGMSMGLRHHSYLGALVALVAAIDDPQFLGAAE
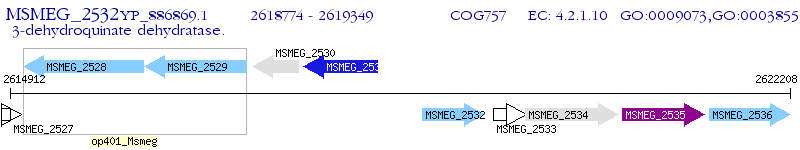
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2532 | aroQ | - | 100% (191) | dehydroquinase dehydratase, type II |
| M. smegmatis MC2 155 | MSMEG_1922 | aroQ | 2e-12 | 28.97% (145) | 3-dehydroquinate dehydratase, type II |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2566c | aroD | 4e-14 | 34.27% (143) | 3-dehydroquinate dehydratase |
| M. gilvum PYR-GCK | Mflv_4683 | - | 3e-16 | 31.97% (147) | 3-dehydroquinate dehydratase, type II |
| M. tuberculosis H37Rv | Rv2537c | aroD | 4e-14 | 34.27% (143) | 3-dehydroquinate dehydratase |
| M. leprae Br4923 | MLBr_00519 | aroD | 2e-13 | 32.39% (142) | 3-dehydroquinate dehydratase |
| M. abscessus ATCC 19977 | MAB_2840c | - | 1e-11 | 30.20% (149) | 3-dehydroquinate dehydratase |
| M. marinum M | MMAR_2178 | aroD | 2e-13 | 33.10% (142) | 3-dehydroquinate dehydratase AroD |
| M. avium 104 | MAV_3413 | aroQ | 4e-13 | 32.87% (143) | 3-dehydroquinate dehydratase |
| M. thermoresistible (build 8) | TH_0107 | aroD | 1e-09 | 31.21% (141) | 3-DEHYDROQUINATE DEHYDRATASE AROD (AROQ) (3-DEHYDROQUINASE) |
| M. ulcerans Agy99 | MUL_1762 | aroD | 2e-13 | 33.10% (142) | 3-dehydroquinate dehydratase |
| M. vanbaalenii PYR-1 | Mvan_1784 | - | 1e-15 | 31.29% (147) | 3-dehydroquinate dehydratase, type II |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2178|M.marinum_M ---------MSEPMTVVN-----------------VLNGPNLGRLGRREP
MUL_1762|M.ulcerans_Agy99 ---------MSEPMTVVN-----------------VLNGPNLGRLGRREP
MAV_3413|M.avium_104 --------------MIVQ-----------------VINGPNLGRLGRREP
Mb2566c|M.bovis_AF2122/97 -----------MSELIVN-----------------VINGPNLGRLGRREP
Rv2537c|M.tuberculosis_H37Rv -----------MSELIVN-----------------VINGPNLGRLGRREP
MLBr_00519|M.leprae_Br4923 -------------MTMVN-----------------IINGPNLGRLGRREP
MAB_2840c|M.abscessus_ATCC_199 -------------MTIQ------------------VLNGPNLGRLGKREA
TH_0107|M.thermoresistible__bu -------------MTAQST--------------VYVINGPNLGRLGRRQP
Mflv_4683|M.gilvum_PYR-GCK -------------MTERR---------------LLLLNGPNLNLLGTRQP
Mvan_1784|M.vanbaalenii_PYR-1 -------------MTERR---------------LLLVNGPNLNLLGTRQP
MSMEG_2532|M.smegmatis_MC2_155 MRRRTVSTYSGEKMTQSSTKVFGWQRTGTRPLLITLIDGPNMPNLGNRNK
:::***: ** *:
MMAR_2178|M.marinum_M DVYG-STTHAELAALIEREAAGLGLQAVVRQSDSEAELLDWIHRAADAGQ
MUL_1762|M.ulcerans_Agy99 DVYG-STTHAELAALIEREAAGLGLQAVVRQSDSEAELLDWIHRAADAGQ
MAV_3413|M.avium_104 DVYG-DTTHDQLAALIEAEAAALGLKAIVRQSDSEAELLDWIHGAADANQ
Mb2566c|M.bovis_AF2122/97 AVYG-GTTHDELVALIEREAAELGLKAVVRQSDSEAQLLDWIHQAADAAE
Rv2537c|M.tuberculosis_H37Rv AVYG-GTTHDELVALIEREAAELGLKAVVRQSDSEAQLLDWIHQAADAAE
MLBr_00519|M.leprae_Br4923 DVYG-STTHEQLSALIERAAVEFGIKAVVRQSDSESQLLDWIHLAADAGE
MAB_2840c|M.abscessus_ATCC_199 AVYG-SVTYAELVSLIEAEAAVLGVDVQVRQSDSEAELLGWVHDAADAKD
TH_0107|M.thermoresistible__bu EVYG-STTHDDLVAMIEREATDLGLKAVVRQSDSEAELLGWIHDAADAGV
Mflv_4683|M.gilvum_PYR-GCK EVYG-STTLAEIEAKVAEVAKDAGLQVRAVQSNHEGVLVDAIHAARTECV
Mvan_1784|M.vanbaalenii_PYR-1 EVYG-ATTLAEIEARVAEVAAEAGLQVRSVQSNHEGVLVDAIHAARTDCA
MSMEG_2532|M.smegmatis_MC2_155 RVYGPISSITALQDHVRELGEGMGVTIEALASNHEGEILDAIHASAARSD
*** : : : . *: *: *. ::. :* :
MMAR_2178|M.marinum_M PVILNAGGLTHTSVALRDACAELSAPLIEVHISNVHAREEFRR-------
MUL_1762|M.ulcerans_Agy99 PVILNAGGLTHTSVALRDACAELSAPLIEVHISNVHAREEFRR-------
MAV_3413|M.avium_104 PVILNAGGLTHTSVALRDACAELSAPLIEVHISNVHAREEFRR-------
Mb2566c|M.bovis_AF2122/97 PVILNAGGLTHTSVALRDACAELSAPLIEVHISNVHAREEFRR-------
Rv2537c|M.tuberculosis_H37Rv PVILNAGGLTHTSVALRDACAELSAPLIEVHISNVHAREEFRR-------
MLBr_00519|M.leprae_Br4923 PVILNAGGLTHTSVALRDACAELSAPFIEVHISNVHAREEFRR-------
MAB_2840c|M.abscessus_ATCC_199 PVILNAGAFTHTSVALRDACAELSAGLIEVHISNVHAREEFRH-------
TH_0107|M.thermoresistible__bu PVILNAGAFTHTSIALRDACAELRAPLIEVHISNVHAREEFRH-------
Mflv_4683|M.gilvum_PYR-GCK GIIINPAAYSHTSVAIADALRSVGLPVAEVHLSNIHAREEFRH-------
Mvan_1784|M.vanbaalenii_PYR-1 GIIINPAAYSHTSVAIADALRSVGLPVAEVHLSNIHAREEFRH-------
MSMEG_2532|M.smegmatis_MC2_155 AYILNPAGLTKTGEPTRHALEETGRPWVEVHFANISAPPWSERGLPGGPW
*:*... ::*. . .* . ***::*: * .:
MMAR_2178|M.marinum_M HSYLSPVATGVIVGLGIQGYLLALRYLASY-----------
MUL_1762|M.ulcerans_Agy99 HSYLSPVATGVIVGLGIRGYLLALRYLASY-----------
MAV_3413|M.avium_104 HSYLSPVATGVIVGLGVQGYLLALRYLAGRPA---------
Mb2566c|M.bovis_AF2122/97 HSYLSPIATGVIVGLGIQGYLLALRYLAEHVGT--------
Rv2537c|M.tuberculosis_H37Rv HSYLSPIATGVIVGLGIQGYLLALRYLAEHVGT--------
MLBr_00519|M.leprae_Br4923 HSYLSPLATGVITGLGVQGYLLALRYLAEIAGN--------
MAB_2840c|M.abscessus_ATCC_199 HSYLSALATGVIVGLGVQGYLLALRYFVASASS--------
TH_0107|M.thermoresistible__bu HSYLSAVATGVIVGLGVQGYLLALRYLANTGKSA-------
Mflv_4683|M.gilvum_PYR-GCK HSYVSAVADMVVAGAGPLGYEMAVQYLADRLGP--------
Mvan_1784|M.vanbaalenii_PYR-1 HSYVSAVADMVVAGAGPLGYEMAVRFLADRLGR--------
MSMEG_2532|M.smegmatis_MC2_155 ESNFTRSATGMSMGLRHHSYLGALVALVAAIDDPQFLGAAE
.* .: * : * .* *: :.