For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VFESLSDRLTGALQGLRGKGRLTDADIDATTREIRLALLEADVSLPVVRAFVARIKERAKGAEVSAALNP AQQVVKIVNEELIGILGGETRQLAFAKTPPTVIMLAGLQGAGKTTLAGKLAKWLKDKGHSPLLVACDLQR PGAVNQLQIVGERAGVAVFAPHPGTAPGAHETEGAGPGDPVAVAAAGLAEAKTKLYGVVIVDTAGRLGID DVLMAQAAGIRDAVQPDETLFVLDAMIGQDAVATAEAFREGVGFTGVVLTKLDGDARGGAALSVREVTGV PILFASAGEKLEDFDVFHPDRMASRILGMGDVLTLIEQAEQVFDAQKAEEAAAKIGTGELTLEDFLEQML TIRKMGPIGNLLGMLPGAGQMKDALAAVDDKQLDRVQAIIRGMTPQERADPKIINASRRLRIANGSGVTV SEVNQLVDRFFEARKMMSSMAGQMGMGFGRKSATRKAAKSKGKKGKNAKKGKGPTQPKVRNPLGAAGLPG GFPDLSNMPKGLDELPPGLADFDLSKLKFPNK
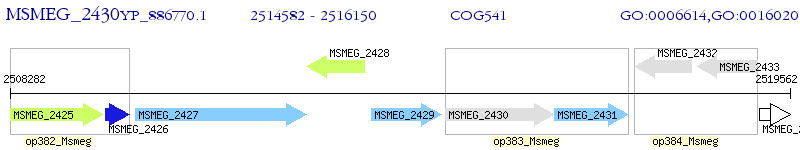
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2430 | ffh | - | 100% (522) | signal recognition particle protein |
| M. smegmatis MC2 155 | MSMEG_2424 | ftsY | 2e-32 | 33.33% (294) | signal recognition particle-docking protein FtsY |
| M. smegmatis MC2 155 | MSMEG_4869 | - | 6e-06 | 27.23% (202) | LAO/AO transport system ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2940c | ffh | 0.0 | 84.91% (530) | signal recognition particle protein |
| M. gilvum PYR-GCK | Mflv_4182 | - | 0.0 | 86.33% (534) | signal recognition particle protein |
| M. tuberculosis H37Rv | Rv2916c | ffh | 0.0 | 84.91% (530) | signal recognition particle protein |
| M. leprae Br4923 | MLBr_01622 | ffh | 0.0 | 80.79% (531) | signal recognition particle protein |
| M. abscessus ATCC 19977 | MAB_3237c | - | 0.0 | 82.13% (526) | Signal recognition particle protein Ffh |
| M. marinum M | MMAR_1792 | ffh | 0.0 | 81.84% (534) | signal recognition particle protein Ffh |
| M. avium 104 | MAV_3770 | ffh | 0.0 | 85.80% (521) | signal recognition particle protein |
| M. thermoresistible (build 8) | TH_2395 | ffh | 0.0 | 87.68% (479) | PROBABLE SIGNAL RECOGNITION PARTICLE PROTEIN FFH (FIFTY-FOUR |
| M. ulcerans Agy99 | MUL_2040 | ffh | 0.0 | 82.77% (528) | signal recognition particle protein Ffh |
| M. vanbaalenii PYR-1 | Mvan_2181 | - | 0.0 | 88.15% (523) | signal recognition particle protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2940c|M.bovis_AF2122/97 ---------------MFESLSDRLTAALQGLRGKGRLTDADIDATTREIR
Rv2916c|M.tuberculosis_H37Rv ---------------MFESLSDRLTAALQGLRGKGRLTDADIDATTREIR
MMAR_1792|M.marinum_M ---------------MFESLSDRLTGALQGLRGKGRLTDADIDATAREIR
MUL_2040|M.ulcerans_Agy99 ---------------MFESLSDRLTGALQGLRGKGRLTDADIDATTREIR
MLBr_01622|M.leprae_Br4923 ---------------MFESLSNRLTGVIQGLRGKGRLTDADIEATTREIR
MAV_3770|M.avium_104 ---------------MFESLSDRLTSALAGLRGKGRLTDADIEATTREIR
MAB_3237c|M.abscessus_ATCC_199 ---------------MFESLSDRLTDALAGLRGKGRLSDADIDATCRTIR
Mflv_4182|M.gilvum_PYR-GCK ---------------MFESLSDRLTGALSGLRGKGRLTDADIDATAREIR
Mvan_2181|M.vanbaalenii_PYR-1 ---------------MFESLSDRLTGALSGLRGKGRLTDADIDATAREIR
MSMEG_2430|M.smegmatis_MC2_155 ---------------MFESLSDRLTGALQGLRGKGRLTDADIDATTREIR
TH_2395|M.thermoresistible__bu MSGPNPPGSDSAGSDRPEPESGRAGDQPDPEQPSDDVADHAPETAETDFY
*. *.* : .. ::* ::: :
Mb2940c|M.bovis_AF2122/97 LALLEA--------------DVSLPVV----RAFIHRIKERARGAEVSSA
Rv2916c|M.tuberculosis_H37Rv LALLEA--------------DVSLPVV----RAFIHRIKERARGAEVSSA
MMAR_1792|M.marinum_M LALLEA--------------DVSLPVV----RAFVHRIKERARGAVVSGA
MUL_2040|M.ulcerans_Agy99 LALLEA--------------DVSLPVV----RAFVHRIKERARGAVVSGA
MLBr_01622|M.leprae_Br4923 LALFEA--------------DVSLPVV----RAFVHRIKERARGTEVSAA
MAV_3770|M.avium_104 LALLEA--------------DVSLPVV----RAFVTRIKERAKGAEVSGA
MAB_3237c|M.abscessus_ATCC_199 LALLEA--------------DVALPVV----RTFITRIKERAKGAEVSGA
Mflv_4182|M.gilvum_PYR-GCK LALLEA--------------DVSLPVV----RAFISRIKDRAKGVEVSAA
Mvan_2181|M.vanbaalenii_PYR-1 LALLEA--------------DVSLPVV----RAFISRIKERAKGAEVSAA
MSMEG_2430|M.smegmatis_MC2_155 LALLEA--------------DVSLPVV----RAFVARIKERAKGAEVSAA
TH_2395|M.thermoresistible__bu SRAYSAPESEQFSSGPYVPPDAALYAXXXGGRNFVARVKERAKGAEVSAA
.* *.:* . * *: *:*:**:*. **.*
Mb2940c|M.bovis_AF2122/97 LNPAQQVVKIVNEELISILGGETRELAFAKTPPTVVMLAGLQGSGKTTLA
Rv2916c|M.tuberculosis_H37Rv LNPAQQVVKIVNEELISILGGETRELAFAKTPPTVVMLAGLQGSGKTTLA
MMAR_1792|M.marinum_M LNPAQQVVKIVNEELVGILGGQTRELVFAKTPPTVVMLAGLQGSGKTTLA
MUL_2040|M.ulcerans_Agy99 LNPAQQVVKIVNEELVGILGGQTRELVFAKTPPTVVMLAGLQGSGKTTLA
MLBr_01622|M.leprae_Br4923 LNPAQQIVKIVNEELIGILGGETRQLAFAKTPPTVVMLAGLQGSGKTTLA
MAV_3770|M.avium_104 LNPAQQVVKIVNEELIGILGGETRQLAFAKTPPTVIMLAGLQGSGKTSLA
MAB_3237c|M.abscessus_ATCC_199 LNPAQQVVKIVNEELIGILGGETRQLAFAKTPPTVVMLAGLQGAGKTTLA
Mflv_4182|M.gilvum_PYR-GCK LNPAQQVVKIVNEELVAILGGETRQLAFARNPPTVIMLAGLQGSGKTTLA
Mvan_2181|M.vanbaalenii_PYR-1 LNPAQQVVKIVNEELIGILGGETRQLAFARNPPTVIMLAGLQGSGKTTLA
MSMEG_2430|M.smegmatis_MC2_155 LNPAQQVVKIVNEELIGILGGETRQLAFAKTPPTVIMLAGLQGAGKTTLA
TH_2395|M.thermoresistible__bu LNPAQQVVKIVNEELIAILGGETRPLKFAKTPPTVIMLAGLQGSGKTTLA
******:********:.****:** * **:.****:*******:***:**
Mb2940c|M.bovis_AF2122/97 GKLAARLRGQGHTPLLVACDLQRPAAVNQLQVVGERAGVPVFAPHPGASP
Rv2916c|M.tuberculosis_H37Rv GKLAARLRGQGHTPLLVACDLQRPAAVNQLQVVGERAGVPVFAPHPGASP
MMAR_1792|M.marinum_M GKLAARLRGQGHTPLLVACDLQRPAAVNQLQVVGQRAGVPVFAPHPGASP
MUL_2040|M.ulcerans_Agy99 GKLAARLRGQGHTPLLVACDLQRPAAVNQLQVVGQRAGVPVFAPHLGASP
MLBr_01622|M.leprae_Br4923 GKLAVRLRRQGHTPLLVACDLQRPAAVNQLQVVGERAGVPVFAPHPGASP
MAV_3770|M.avium_104 GKLAAWLRGQGHTPLLVACDLQRPAAVNQLQVVGERAGVPVFAPHPGASP
MAB_3237c|M.abscessus_ATCC_199 GKLAKWLKGQGHTPLLVACDLQRPGAVNQLQVVGERAGVSVYAPHPGTAD
Mflv_4182|M.gilvum_PYR-GCK GKLAKWLKAQGHTPLLVACDLQRPGAVNQLQIVGERAGVHVFAPHPGVSP
Mvan_2181|M.vanbaalenii_PYR-1 GKLAKWLKGQGHTPLLVACDLQRPGAVNQLQIVGERAGVSVFAPHPGVSP
MSMEG_2430|M.smegmatis_MC2_155 GKLAKWLKDKGHSPLLVACDLQRPGAVNQLQIVGERAGVAVFAPHPGTAP
TH_2395|M.thermoresistible__bu GKLAKWLRDKGHTPLLVACDLQRPGAVNQLQIVGERAGVAVFAPHPGVSA
**** *: :**:***********.******:**:**** *:*** *.:
Mb2940c|M.bovis_AF2122/97 -----ESG----------PGDPVAVAAAGLAEARAKHFDVVIVDTAGRLG
Rv2916c|M.tuberculosis_H37Rv -----ESG----------PGDPVAVAAAGLAEARAKHFDVVIVDTAGRLG
MMAR_1792|M.marinum_M -----ESG----------PGDPVAVAAAGLAEAQAKHFDVVIVDTAGRLG
MUL_2040|M.ulcerans_Agy99 -----ESG----------PGDPVAVAAAGLAEAQAKHFDVVIVDTAGRLG
MLBr_01622|M.leprae_Br4923 -----DSG----------PGDPVAVASAGLVEARAKHFDVVIVDTAGRLG
MAV_3770|M.avium_104 -----ESG----------PGDPVAVAAQGLAEARAKHHDVVIVDTAGRLG
MAB_3237c|M.abscessus_ATCC_199 G--GLEVGSL--------VGDPVAVARDGLAHAKAQHFDVVLVDTAGRLG
Mflv_4182|M.gilvum_PYR-GCK AGETIEQMTAHARSETAGAGDPVSVAAAGLAEARSKLYDVVIVDTAGRLG
Mvan_2181|M.vanbaalenii_PYR-1 DGATIEQMTS---------GDPVSVAAAGLAEARSKHYDVVIVDTAGRLG
MSMEG_2430|M.smegmatis_MC2_155 GAHETEGAG---------PGDPVAVAAAGLAEAKTKLYGVVIVDTAGRLG
TH_2395|M.thermoresistible__bu SG-AAEDG----------PGDPVAVAAAGVAEAKAKLFDVVIVDTAGRLG
: ****:** *:..*::: ..**:********
Mb2940c|M.bovis_AF2122/97 IDEELMAQAAAIRDAINPDEVLFVLDAMIGQDAVTTAAAFGEGVGFTGVA
Rv2916c|M.tuberculosis_H37Rv IDEELMAQAAAIRDAINPDEVLFVLDAMIGQDAVTTAAAFGEGVGFTGVA
MMAR_1792|M.marinum_M IDEELMAQASAIRKAINPDEVLFVLDAMIGQDAVATAQAFGEGVGFTGVA
MUL_2040|M.ulcerans_Agy99 IDEELMAQASAIRKAINPDEVLFVLDAMIGQDAVATAQAFGEGVGFTGVA
MLBr_01622|M.leprae_Br4923 IDDELMAQAGAIREAINPDEVLFVLDAMIGQDAVTTAAAFGAGVGFTGVV
MAV_3770|M.avium_104 IDDELMAQAAAIRDAIDPDEILFVLDAMIGQDAVTTAEAFREGVGFTGVV
MAB_3237c|M.abscessus_ATCC_199 IDAELMDQAARIRDAVDPDEVLFVLDAMIGQDAVSTAEAFREGVGFTGVV
Mflv_4182|M.gilvum_PYR-GCK IDQELMAQAAAIRDAVNPDEILFVLDAMIGQDAVTTADAFREGVGFTGVV
Mvan_2181|M.vanbaalenii_PYR-1 IDAELMAQAAAIRDAVNPDEVLFVLDAMIGQDAVTTADAFREGVGFTGVV
MSMEG_2430|M.smegmatis_MC2_155 IDDVLMAQAAGIRDAVQPDETLFVLDAMIGQDAVATAEAFREGVGFTGVV
TH_2395|M.thermoresistible__bu IDDELMNQAAAIRDAVDPDEVLFVLDAMIGQDAVTTAEAFREGVGFTGVV
** ** **. **.*::*** *************:** ** *******.
Mb2940c|M.bovis_AF2122/97 LTKLDGDARGGAALSVREVTGVPILFASTGEKLEDFDVFHPDRMASRILG
Rv2916c|M.tuberculosis_H37Rv LTKLDGDARGGAALSVREVTGVPILFASTGEKLEDFDVFHPDRMASRILG
MMAR_1792|M.marinum_M LTKLDGDARGGAALSVREVTGVPILFASTGEKLEDFDVFHPDRMASRILG
MUL_2040|M.ulcerans_Agy99 LTKLDGDARGGAALSVREVTGVPILFASTGEKLEDFDVFHPDRMASRILG
MLBr_01622|M.leprae_Br4923 LTKLDGDARGGAALSVREVTGVPILFATTGEKLDDFDVFHPDRMASRILG
MAV_3770|M.avium_104 LTKLDGDARGGAALSVREVTGVPILFASTGEKLEDFDVFHPDRMSSRILG
MAB_3237c|M.abscessus_ATCC_199 LTKLDGDARGGAALSVREITGTPILFASTGEKLEDFDVFHPDRMASRILG
Mflv_4182|M.gilvum_PYR-GCK LTKLDGDARGGAALSVREVTGQPILFASAGEKLEDFDVFHPDRMASRILG
Mvan_2181|M.vanbaalenii_PYR-1 LTKLDGDARGGAALSVREVTGVPILFASAGEKLEDFDVFHPDRMASRILG
MSMEG_2430|M.smegmatis_MC2_155 LTKLDGDARGGAALSVREVTGVPILFASAGEKLEDFDVFHPDRMASRILG
TH_2395|M.thermoresistible__bu LTKLDGDARGGAALSVREVTGVPILFASAGEKLEDFDVFHPDRMAGRILG
******************:** *****::****:**********:.****
Mb2940c|M.bovis_AF2122/97 MGDVLSLIEQAEQVFDAQQAEEAAAKIGAGELTLEDFLEQMLAVRKMGPI
Rv2916c|M.tuberculosis_H37Rv MGDVLSLIEQAEQVFDAQQAEEAAAKIGAGELTLEDFLEQMLAVRKMGPI
MMAR_1792|M.marinum_M MGDVLSLIEQAEQVFDAEQAEQAAAKIGAGELTLEDFLEQMLAVRKMGPI
MUL_2040|M.ulcerans_Agy99 MGDVLSLIEQAEQVFDAEQAEQAAAKIGAGELTLEDFLEQMLAVRKMGPI
MLBr_01622|M.leprae_Br4923 MGDVLSLIEQAEQVFDAEQAEAAAAKIVAGELTLEDFLEQMLAVRKMGLI
MAV_3770|M.avium_104 MGDVLSLIEQAEQVFDAEQAEAAAVKIGSGELTLEDFLEQMLAIRKMGPI
MAB_3237c|M.abscessus_ATCC_199 MGDLLSLIEQAEQHFDAEQSLAAAEKITSGELTLEDFLEQMLAIRKMGPI
Mflv_4182|M.gilvum_PYR-GCK MGDVLTLIEQAEQVFDAEQAEATAAKIGSGELTLEDFLEQMLAIRKMGPI
Mvan_2181|M.vanbaalenii_PYR-1 MGDVLTLIEQAEQHFDAAQAEATAAKIGSGELTLEDFLEQMLAIRKMGPI
MSMEG_2430|M.smegmatis_MC2_155 MGDVLTLIEQAEQVFDAQKAEEAAAKIGTGELTLEDFLEQMLTIRKMGPI
TH_2395|M.thermoresistible__bu MGDVLTLIEQAEQVFDAQQAEEAAVKIGTGELTLEDFLEQMQAIRKMGPI
***:*:******* *** :: :* ** :************ ::**** *
Mb2940c|M.bovis_AF2122/97 GNLLGMLPGAAQMKDALAEVDDKQLDRVQAIIRGMTPQERADPKIINASR
Rv2916c|M.tuberculosis_H37Rv GNLLGMLPGAAQMKDALAEVDDKQLDRVQAIIRGMTPQERADPKIINASR
MMAR_1792|M.marinum_M GNLLGMLPGGAQMKDALAEVDDKHLDRIQAIIRGMTPEERADPKIINASR
MUL_2040|M.ulcerans_Agy99 GNLLGMLPGGAQMKDALAEVDDKHLDRIQAIIRGMTPEERADPKIINASR
MLBr_01622|M.leprae_Br4923 GNLLGMLPGAGQVKEVLEQVDDKQLDRLQAIIRGMTPQERADPKIINASR
MAV_3770|M.avium_104 GNLLGMLPGAGQMKDALAAVDDKQLDRLQAIIRGMTPEERADPKIINASR
MAB_3237c|M.abscessus_ATCC_199 GNLLGMLPGAGQMKDALAAVDDKQLDRVQAIIRGMTPAERADPKIINASR
Mflv_4182|M.gilvum_PYR-GCK GNLLGMLPGAGQMKDALAAVDDSQLDRVQAIIRGMTPQERADPKIINASR
Mvan_2181|M.vanbaalenii_PYR-1 GNLLGMLPGAGQMKDALAAVDDKQLDRVQAIIRGMTPQEREDPKIINASR
MSMEG_2430|M.smegmatis_MC2_155 GNLLGMLPGAGQMKDALAAVDDKQLDRVQAIIRGMTPQERADPKIINASR
TH_2395|M.thermoresistible__bu GNLLGMLPGAGQMKDALAAVDDKQLDRVQAIIRGMTPEERADPKIINASR
*********..*:*:.* ***.:***:********* ** *********
Mb2940c|M.bovis_AF2122/97 RLRIANGSGVTVSEVNQLVERFFEARKMMSSMLGGMGIPGIGRKSATRKS
Rv2916c|M.tuberculosis_H37Rv RLRIANGSGVTVSEVNQLVERFFEARKMMSSMLGGMGIPGIGRKSATRKS
MMAR_1792|M.marinum_M RLRIANGSGVSVSEVNQLVDRFFEARKMMSSMLGGMGIPGMGRKSATRKS
MUL_2040|M.ulcerans_Agy99 RLRIANGSGVSVSEVNQLVDRFFEARKMMSSMLGGMGIPGMGRKSATRKS
MLBr_01622|M.leprae_Br4923 RLRIANGSGVTVSEVNQLVDRFFEARKMMSSVLGGMGIPGMGRKSATRKS
MAV_3770|M.avium_104 RLRIANGSGVTVSEVNQLVDRFFEARKMMSSMLGGMGIPGLGRKSATRKA
MAB_3237c|M.abscessus_ATCC_199 RLRIANGSGVAVSEVNQLVDRFFEARKMMSSMAGRMGMGAINRKSNR---
Mflv_4182|M.gilvum_PYR-GCK RLRIANGSGVTVAEVNQLVDRFFEARKMMSQMAGQMGMP-FGRKNTR---
Mvan_2181|M.vanbaalenii_PYR-1 RLRIANGSGVTVSEVNQLVDRFFEARKMMSQMAGQMGMP-FGRKNTR---
MSMEG_2430|M.smegmatis_MC2_155 RLRIANGSGVTVSEVNQLVDRFFEARKMMSSMAGQMGMG-FGRKSATRKA
TH_2395|M.thermoresistible__bu RRRIAEGSGVTVAEVNQLVERFFEARKMMSQMAGQMGIPGIGRKNTR--K
* ***:****:*:******:**********.: * **: :.**.
Mb2940c|M.bovis_AF2122/97 KGAKGKSGKKSKKG-TRGPTPPKVKSPFG--VPGMPG---LAGLPGGLPD
Rv2916c|M.tuberculosis_H37Rv KGAKGKSGKKSKKG-TRGPTPPKVKSPFG--VPGMPG---LAGLPGGLPD
MMAR_1792|M.marinum_M KGAKGKAGKKGKKGGARGPTPPKVKSPFG--MPGMPGMPGMPGMPAGFPD
MUL_2040|M.ulcerans_Agy99 KGAKGKAGKKGKKGGARGPTPPKVKSPFG--MPGMP------GMLAGFPD
MLBr_01622|M.leprae_Br4923 KGGKGK--KR-----ARGPTSPKVRSPFGPARPGMPD---MMNMPPSFPD
MAV_3770|M.avium_104 KGAKGK-AKKGKKG-ARGPTPPKVRSPLG---PGMP---------AGYPD
MAB_3237c|M.abscessus_ATCC_199 KGGKNN--KKGKKG-SRGPTAPKLKGGFPG-MAGLPG-----GMPAGFPD
Mflv_4182|M.gilvum_PYR-GCK KAAKGKNKQAGKKKGGRGPTPPKNRNPLG---AGMPG---MAGLPPGFPD
Mvan_2181|M.vanbaalenii_PYR-1 KAAKGKNKQAGKK-GRKGPTPPKNRNPLG---AGMP------GMPAGFPD
MSMEG_2430|M.smegmatis_MC2_155 AKSKGKKGKNAKKG--KGPTQPKVRNPLG--AAGLPG---------GFPD
TH_2395|M.thermoresistible__bu AGKKGKKGQRGRKK-GAGPTPPKVRNPLG---APMPG------MPAGFPD
*.: : *** ** :. : . :* . **
Mb2940c|M.bovis_AF2122/97 LSQMPKGLDELPPGLADFDLSKLKFPGKK
Rv2916c|M.tuberculosis_H37Rv LSQMPKGLDELPPGLADFDLSKLKFPGKK
MMAR_1792|M.marinum_M LSQMPEGLDELPPGLADFDLSKLNFPGKK
MUL_2040|M.ulcerans_Agy99 LSQMPEGLDELPPGLADFDLSKLNFPGKK
MLBr_01622|M.leprae_Br4923 LSQMPDGLNELPPGLAAFDLSKLKFPGKT
MAV_3770|M.avium_104 LSQLPEGLNELPPGLADFDLSKLKFPGNK
MAB_3237c|M.abscessus_ATCC_199 LSGMPEGLGELPPGLADIDISKLKFPKN-
Mflv_4182|M.gilvum_PYR-GCK LSNMPKGLDELPPGLADIDLSKLKFPKN-
Mvan_2181|M.vanbaalenii_PYR-1 LSNMPKGLDELPPGLADIDLSKLKFPKN-
MSMEG_2430|M.smegmatis_MC2_155 LSNMPKGLDELPPGLADFDLSKLKFPNK-
TH_2395|M.thermoresistible__bu LSQMPPGLDELPPGLADIDLSKLKFPGKK
** :* **.******* :*:***:** :