For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MCLGIPGKVVAVWDEAGTRMSTVDFGGTTKTVCLAYLPDMQIGEYTIVHAGFAITRLDEASAQETLRMFK DLGVLDEELAEGVETA
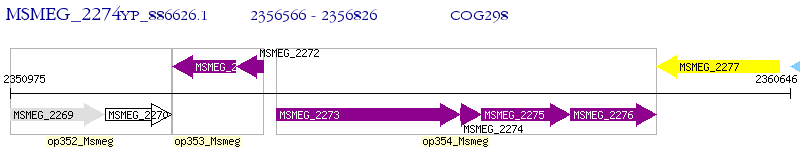
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2274 | hypC | - | 100% (86) | hydrogenase assembly chaperone HypC/HupF |
| M. smegmatis MC2 155 | MSMEG_2712 | hypC | 2e-09 | 36.00% (75) | hydrogenase assembly chaperone HypC/HupF |
| M. smegmatis MC2 155 | MSMEG_2703 | hypC | 7e-09 | 34.18% (79) | hydrogenase assembly chaperone HypC/HupF |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3980 | - | 2e-26 | 58.14% (86) | hydrogenase assembly chaperone hypC/hupF |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1863 | - | 4e-09 | 34.67% (75) | HupF/HypC family protein |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0222 | hypC | 4e-23 | 57.69% (78) | hydrogenase assembly chaperone HypC/HupF |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2360 | - | 6e-33 | 86.11% (72) | hydrogenase assembly chaperone hypC/hupF |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2274|M.smegmatis_MC2_155 MCLGIPGKVVAVWDEA-GTRMSTVDFGGTTKTVCLAYLPDMQI--GEYTI
Mvan_2360|M.vanbaalenii_PYR-1 --------MIEIWEEA-GTRMSTVEFGGTTKTVCLAYLPDMEV--GEYTI
Mflv_3980|M.gilvum_PYR-GCK MCLAVPGRILSIHERD-GTTMSVVDFGGIRKNVCLQYLPDAQV--GQYVV
TH_0222|M.thermoresistible__bu MCLGIPGRIVEIWDEDSGARMARVAFPGEERTACLAYLPDLQV--GDYTI
MMAR_1863|M.marinum_M MCLGIPGQVIRILDGY-EGQLALVEVTGEARKVNIGMLPDETFSPGDWVI
:: : : :: * . * :.. : *** . *::.:
MSMEG_2274|M.smegmatis_MC2_155 VHAGFAITRLDEASAQETLRMFKDLG--VLDEELAEG-----VETA----
Mvan_2360|M.vanbaalenii_PYR-1 VHAGFAIARLDEASAHETLRMFADLG--VLDEELAGEQPVGRRDPA----
Mflv_3980|M.gilvum_PYR-GCK VHVGFAIQRLDEESARRTLAEFAHLG--VLDEEFADGFASAARQARLANP
TH_0222|M.thermoresistible__bu LHAGFALTRLDEDQAELTLATMAEYG--VFGDGSAVTA------------
MMAR_1863|M.marinum_M IHMGFAIEKTDRAGAEQAMAGLELMGRGRTDTDFTDGG------------
:* ***: : *. *. :: : * . :
MSMEG_2274|M.smegmatis_MC2_155 ----------
Mvan_2360|M.vanbaalenii_PYR-1 ----------
Mflv_3980|M.gilvum_PYR-GCK DTGARSESIP
TH_0222|M.thermoresistible__bu ----------
MMAR_1863|M.marinum_M ----------