For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QRHIDAATSVAGLRAATAELPAAARELVGARAATVAHAWSALLRAALRTAARVTGPAPGTWYASGSVGRG EALPGSDLDTLFVRDEGTPVDAALHHALEVHDLLSACGIPGDEKGVTAARARFNRSASQWEDGIARWAGD AAADRGVVMIGLLADAFPLTDGPDLAARARTAVGDHPDALAAMLQDATYLRAHVPSRLRVLATRDTAVDL KAATIDPVVRITRWAALCCGSSERGTPDRIHDAAGGRFLDDEDAAVLTRCHAIGSSIRWRTRADQITSSG GEITDHVAMSAFSPQDRTALRSIGRDVSGLQRKLDYLASTSTFSPW*
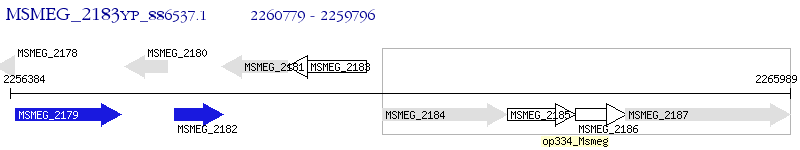
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2183 | - | - | 100% (327) | hypothetical protein MSMEG_2183 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3086 | - | 2e-28 | 30.24% (334) | cyclic nucleotide-binding protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2708 | - | 1e-61 | 43.41% (334) | hypothetical protein MAB_2708 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2183|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2708|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3086|M.gilvum_PYR-GCK MPSSSGPAEPADLPTFLAAHTPFQGSTAEELAELAVGAQLETFPAAAVVA
MSMEG_2183|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2708|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3086|M.gilvum_PYR-GCK DYSARVPDEVWMVRDGAVALRSSGDGALIDVVTAGGIFGYLPLLTGAGAD
MSMEG_2183|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2708|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3086|M.gilvum_PYR-GCK FEARTTSPSTLVRLPGAAVRAQFAKPSGLAFLASAGWNRPADRPTIAPAT
MSMEG_2183|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2708|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3086|M.gilvum_PYR-GCK DSRPVAELVAGGVLLVAPGDPVRDVVVQMTDHHVSYALIRLPDGRLGIFT
MSMEG_2183|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2708|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3086|M.gilvum_PYR-GCK DRDLRIRVVAAGLPVDVAVDRVMSAPARTVTADMTADSVLMEMLECGLRH
MSMEG_2183|M.smegmatis_MC2_155 -----------------------------MQR---HIDAATSVAGLRAAT
MAB_2708|M.abscessus_ATCC_1997 -------------------MSAEVPAENGMARAIAQIDAAADEAAFHAGV
Mflv_3086|M.gilvum_PYR-GCK MPVLTPRGEVIGVLEDADLLAATARQSFLLRRSIGQASDAAQLQQVGQRV
: * : . *:. . .
MSMEG_2183|M.smegmatis_MC2_155 AELPAAARELVG-ARA--ATVAHAWSALLRAALRTAARVT--GPAPG-TW
MAB_2708|M.abscessus_ATCC_1997 SATRIAITLQAR-ARAPEATLAAAWSEALRSSVITAARLVTDGVNPGWSW
Mflv_3086|M.gilvum_PYR-GCK TATAVDLFRNGTNAAATSAILSVVIDSLVRRSLELVLAQPENAGVGGFAW
: * * * :: . . :* :: . . * :*
MSMEG_2183|M.smegmatis_MC2_155 YASGSVGRGEALPGSDLDTLFVRDEGTPVD---AALHHALEVHDLLSACG
MAB_2708|M.abscessus_ATCC_1997 FVSGSAARGEAAPGSDIETLIAVDDTIDAEGKTRLMELAARVHTTLERGG
Mflv_3086|M.gilvum_PYR-GCK LTLGSVARREAMPSSDVDSALSWRDDLDGEAP-RLRGIAADVHHILDGCG
. **..* ** *.**::: : : : * ** *. *
MSMEG_2183|M.smegmatis_MC2_155 IPGDEKGVTAARARFNRSASQWEDGIARWAGDAAADRGVVMIGLLADA--
MAB_2708|M.abscessus_ATCC_1997 IGGDSNGVLASRGRFCRRTTDWLEGIERWCANPPADRGVVMAGLMADSRG
Mflv_3086|M.gilvum_PYR-GCK LPADSNGAIATTTAFARSQSDWRRAAQGWVDEPLRNLGLIMSSLLLDARV
: .*.:*. *: * * ::* . * :. : *::* .*: *:
MSMEG_2183|M.smegmatis_MC2_155 ----FPLTDGPDLAARARTAVGDHPDALAAMLQDATYLRAHVPSRLRVLA
MAB_2708|M.abscessus_ATCC_1997 VGADASLPDGALRSKNARCAQRHYP-IRHAMLQDAVAVRAGFPSRLKIFA
Mflv_3086|M.gilvum_PYR-GCK VWGEVGLHTAPAAYQRIREKN---PEALRLQLLDALAGKMRTRSLRDVLS
* .. . * * * ** : * :::
MSMEG_2183|M.smegmatis_MC2_155 TRDTAVDLKAATIDPVVRITRWAALCCGSSERGTPDRIHDAAGGRFLDDE
MAB_2708|M.abscessus_ATCC_1997 TQSDSVDLKITAMDPVVKIARWAALSAGSDVLATPARLDAASTAETLDRD
Mflv_3086|M.gilvum_PYR-GCK RRGGTFDLKNHALAPIVNLARWGGLSAGVTSASTPARLAAAAQAGVLRDA
:. :.*** :: *:*.::**..*..* .** *: *: . *
MSMEG_2183|M.smegmatis_MC2_155 DAAVLTRCHAIGSSIRWRTRADQITSSGGEITDHVAMSAFSPQDRTALRS
MAB_2708|M.abscessus_ATCC_1997 DASTLHDCFDWLLRFRWRARAAAWEE-GQPMSDTVSLSALAPQERAMLRG
Mflv_3086|M.gilvum_PYR-GCK DSRTLCDVFAMVQRLRLAHQVEQISA-GHPPGDIITMSELSPLNRSLLGD
*: .* . :* :. * * :::* ::* :*: * .
MSMEG_2183|M.smegmatis_MC2_155 IGRDVSGLQRKLDYLASTSTFSPW
MAB_2708|M.abscessus_ATCC_1997 IAREIAGIRRKLVYLSSTSSFR--
Mflv_3086|M.gilvum_PYR-GCK GLREIAAVRRRVGSAGLTGI----
*:::.::*:: . *.