For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STVADDLTLALELADQADALTMDRFGALDLRVETKPDLTPVTDADRGAEETLRAALAQARPADTVFGEEF GGTTASTGRQWVIDPIDGTKNFVRGVPVWCTLIALLDDGVPRVGVVSAPALARRWWAAEGQGAFGSFNGT TRKLSVSGVSDLSAASLSYSDLTTGWDDRRERFVELTDAVWRVRAYGDFWSYCMVAEGAVDIACEPEVKL WDIAPLDVLIREAGGTFTSIDGADGPHGGSALATNGVLHDAVVSRLAVR*
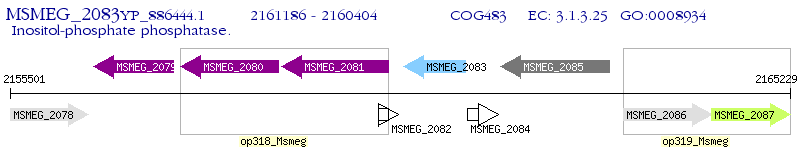
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2083 | - | - | 100% (260) | inositol monophosphatase |
| M. smegmatis MC2 155 | MSMEG_2762 | - | 3e-23 | 35.81% (229) | inositol-1-monophosphatase |
| M. smegmatis MC2 155 | MSMEG_3116 | - | 7e-23 | 33.62% (235) | inositol-1-monophosphatase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3161 | - | 1e-100 | 69.02% (255) | monophosphatase |
| M. gilvum PYR-GCK | Mflv_4454 | - | 1e-119 | 78.76% (259) | histidinol-phosphate phosphatase, putative |
| M. tuberculosis H37Rv | Rv3137 | - | 1e-100 | 69.02% (255) | monophosphatase |
| M. leprae Br4923 | MLBr_00662 | - | 1e-98 | 66.80% (250) | putative monophosphatase |
| M. abscessus ATCC 19977 | MAB_3485 | - | 1e-97 | 66.41% (259) | monophosphatase |
| M. marinum M | MMAR_1511 | - | 1e-100 | 68.50% (254) | monophosphatase |
| M. avium 104 | MAV_4017 | - | 1e-100 | 70.43% (257) | inositol monophosphatase |
| M. thermoresistible (build 8) | TH_2149 | - | 1e-100 | 69.80% (255) | PROBABLE MONOPHOSPHATASE |
| M. ulcerans Agy99 | MUL_2430 | - | 4e-99 | 67.72% (254) | monophosphatase |
| M. vanbaalenii PYR-1 | Mvan_1908 | - | 1e-116 | 75.97% (258) | histidinol-phosphate phosphatase, putative |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1511|M.marinum_M ----------MSEQDLHLALALADRADAVTRARFQALDLRVDTKPDLTPV
MUL_2430|M.ulcerans_Agy99 ----------MSEQDLHLALALADRADAVTRARFQALDLRVDTKPDLTPV
Mb3161|M.bovis_AF2122/97 ----------MSHDDLMLALALADRADELTRVRFGALDLRIDTKPDLTPV
Rv3137|M.tuberculosis_H37Rv ----------MSHDDLMLALALADRADELTRVRFGALDLRIDTKPDLTPV
MLBr_00662|M.leprae_Br4923 ----------------MLALTLADRADALTSARFGALNLRVDTKPDLTPV
MAV_4017|M.avium_104 ----------MSNDDLALALLLADRADAVTSARFGALDLRIDTKPDLTPV
TH_2149|M.thermoresistible__bu MTAVPDDAPDIRDDDLALALLLADQADVVTRDRFGSLDLHVETKPDLTPV
MAB_3485|M.abscessus_ATCC_1997 -----MTPTVSLREDRRLALELADAADEITTSRFGALDLRVDTKPDLTPV
Mflv_4454|M.gilvum_PYR-GCK --------MSTLADDVTLALQLADEADVLTMQRFGAVDLRVETKPDLTPA
Mvan_1908|M.vanbaalenii_PYR-1 --------MSTLVDDLNLALRLADEADALTMQRFGAVDLRVETKPDMTPA
MSMEG_2083|M.smegmatis_MC2_155 --------MSTVADDLTLALELADQADALTMDRFGALDLRVETKPDLTPV
*** *** ** :* ** :::*:::****:**.
MMAR_1511|M.marinum_M TDADRAVETEIRAALQRERPGDSVLGEEYGGTTTLRGRQWIIDPIDGTKN
MUL_2430|M.ulcerans_Agy99 TDADRAVETEIRAALQRERPGDSVLGEEYGGTTTLRGRQWIIDPIDGTKN
Mb3161|M.bovis_AF2122/97 TDADRAVESDVRQTLGRDRPGDGVLGEEFGGSTTFTGRQWIVDPIDGTKN
Rv3137|M.tuberculosis_H37Rv TDADRAVESDVRQTLGRDRPGDGVLGEEFGGSTTFTGRQWIVDPIDGTKN
MLBr_00662|M.leprae_Br4923 TDADRAVEADVRAVLGRERPKDGILGEEYGGTITFSGQQWIVDPIDGTKN
MAV_4017|M.avium_104 TDADRAAEAELRAVLARQRPHDSIVGEEFGGDTAFTGRQWIIDPIDGTKN
TH_2149|M.thermoresistible__bu SDADRSVERILRGTLAQQRPDDPILGEEFGGATVFSGRQWIIDPIDGTKN
MAB_3485|M.abscessus_ATCC_1997 TDADTSVETVLRSLLHRSRPADSVLGEEYGGEATFTGRQWVIDPIDGTKN
Mflv_4454|M.gilvum_PYR-GCK TDADLDAERLLRARLGEARPQDSVFGEEFGGTKEFTGRQWVVDPIDGTKN
Mvan_1908|M.vanbaalenii_PYR-1 TDADLDAEKLLRARLTESRPQDSVFGEEFGGTREFTGRQWVVDPIDGTKN
MSMEG_2083|M.smegmatis_MC2_155 TDADRGAEETLRAALAQARPADTVFGEEFGGTTASTGRQWVIDPIDGTKN
:*** .* :* * . ** * :.***:** *:**::********
MMAR_1511|M.marinum_M FVRGVPVWASLIALLEDGVPSIGVVSAPALQRRWWAGRGQGAFTAVQGGP
MUL_2430|M.ulcerans_Agy99 FVRGVPVWASLIALLEDGVPSIGVVSAPALQRRWWAGRGQGAFTAVQGGP
Mb3161|M.bovis_AF2122/97 FVRGVPVWASLIALLEDGVPSVGVVSAPALQRRWWAARGRGAFASVDGAR
Rv3137|M.tuberculosis_H37Rv FVRGVPVWASLIALLEDGVPSVGVVSAPALQRRWWAARGRGAFASVDGAR
MLBr_00662|M.leprae_Br4923 FVRGVPVWASLIALLEDGVPSIGVVSAPALQRRWWAARGQGAFVAVDGV-
MAV_4017|M.avium_104 FVRGVPVWATLIALLHDGVPVVGAVSAPALARRWWAAHGRGAFVAVNGAA
TH_2149|M.thermoresistible__bu FVRGVPVWATLIALLQDGVPTVGVVSAPALQRRWYAAAGQGAHVVVGDDV
MAB_3485|M.abscessus_ATCC_1997 FARGVPIWATLIALLQDGVPVVGVVSAPALSRRWWAAEGLGAHSRVGSEE
Mflv_4454|M.gilvum_PYR-GCK FVRGVPVWSTLIALLVDGVPVVGVVSAPALGRRWWAGQGQGAFTSFAGT-
Mvan_1908|M.vanbaalenii_PYR-1 FVRGVPVWSTLIALLVDGVPVVGVISAPALGRRWWAGEGQGAFTSFGGD-
MSMEG_2083|M.smegmatis_MC2_155 FVRGVPVWCTLIALLDDGVPRVGVVSAPALARRWWAAEGQGAFGSFNGT-
*.****:*.:***** **** :*.:***** ***:*. * **. . .
MMAR_1511|M.marinum_M PRRIAVSSVAELNSASLSFSSLS-GWAQLGLRGQFVDLTDAVWRVRAYGD
MUL_2430|M.ulcerans_Agy99 PRRIAVSSVAELNSASLSFSSLS-GWAQLGLRGQFVDLTDAVWRVRAYGD
Mb3161|M.bovis_AF2122/97 PHRLSVSSVAELHSASLSFSSLS-GWARLGLRERFIGLTDTVWRVRAYGD
Rv3137|M.tuberculosis_H37Rv PHRLSVSSVAELHSASLSFSSLS-GWARPGLRERFIGLTDTVWRVRAYGD
MLBr_00662|M.leprae_Br4923 PRRLAVSEVADLNSASLSFSSLS-GWAQRGLRDRFLELTDAVWRVRAYGD
MAV_4017|M.avium_104 ARRLSVSSVAQLDSASLSFSSLS-GWAQLGLRDRFLALTDAVWRVRAFGD
TH_2149|M.thermoresistible__bu PRRISVSSVSDLESASLSFSSLS-GWAVRKLRDRFIDLTDTVWRVRGFGD
MAB_3485|M.abscessus_ATCC_1997 ERSIHVSAVDQLSSASLSFSSLS-GWADRGKRDRFIDLTDEIWRVRGFGD
Mflv_4454|M.gilvum_PYR-GCK TRRISVSGVGDLASASLSYSDLTTGWDDK--RARFVELLDAVWRVRGYGD
Mvan_1908|M.vanbaalenii_PYR-1 TRRISVSAVADMEAASLSFSDLATGWEEK--RSRFVELLDAVWRMRGYGD
MSMEG_2083|M.smegmatis_MC2_155 TRKLSVSGVSDLSAASLSYSDLTTGWDDR--RERFVELTDAVWRVRAYGD
: : ** * :: :****:*.*: ** * :*: * * :**:*.:**
MMAR_1511|M.marinum_M FFSYCLLAEGAIDIAAEPEVSVWDLAPLDILVREAGGQFTSLDGTPGPHG
MUL_2430|M.ulcerans_Agy99 FFSYCLLAEGAIDIAAEPEVSVWDLAPLDILVREAGGQFSSLDGTPGPHG
Mb3161|M.bovis_AF2122/97 FLSYCLVAEGAVDIAAEPQVSVWDLAALDIVVREAGGRLTSLDGVAGPHG
Rv3137|M.tuberculosis_H37Rv FLSYCLVAEGAVDIAAEPQVSVWDLAALDIVVREAGGRLTSLDGVAGPHG
MLBr_00662|M.leprae_Br4923 FLSYCLLAEGAIDVAAEPKVSVWDLAALDIVVREAGGVLTGLDGTPGPHG
MAV_4017|M.avium_104 FLSYCLVAEGAVDIAAEPEVSVWDLAALDILVREAGGTLTGLDGTPGPHR
TH_2149|M.thermoresistible__bu FFSYCLLAEGAVDIATEPEVSLWDLAALDILVREAGGTFTNLEGAPGPHG
MAB_3485|M.abscessus_ATCC_1997 FFSYCLVAEGAVDIAAEPEVSLWDLAPLDILVREAGGRFTNLAGEPGPHG
Mflv_4454|M.gilvum_PYR-GCK FWSYCLVAEGAVDIAVEPEVKLWDLAPLDVLVREAGGRFTDLSGGAGPHG
Mvan_1908|M.vanbaalenii_PYR-1 FWSYCLVAEGAVDIAVEPEVKLWDLAPLDILVREAGGRFTNLAGEPGPHG
MSMEG_2083|M.smegmatis_MC2_155 FWSYCMVAEGAVDIACEPEVKLWDIAPLDVLIREAGGTFTSIDGADGPHG
* ***::****:*:* **:*.:**:*.**:::***** ::.: * ***
MMAR_1511|M.marinum_M GSAVASNGLLHEQVLNRLRSDRV
MUL_2430|M.ulcerans_Agy99 GSAVVSNGLLHEQVLNRLRSDRV
Mb3161|M.bovis_AF2122/97 GSAVATNGLLHDEVLTRLNAG--
Rv3137|M.tuberculosis_H37Rv GSAVATNGLLHDEVLTRLNAG--
MLBr_00662|M.leprae_Br4923 GSAVATNGRLHQEVLTRIGATVK
MAV_4017|M.avium_104 GSAVASNGLLQRQVLERLAL---
TH_2149|M.thermoresistible__bu GSAVATNGRLHNAVLAHLRGV--
MAB_3485|M.abscessus_ATCC_1997 GSAVATNGLLHEEVLKALS----
Mflv_4454|M.gilvum_PYR-GCK GSAVATNGRVHDAVLAALDV---
Mvan_1908|M.vanbaalenii_PYR-1 GSAVATNGLLHDAVLSALAD---
MSMEG_2083|M.smegmatis_MC2_155 GSALATNGVLHDAVVSRLAVR--
***:.:** :: *: :