For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGSAPLRILVYSDNPRTREQVRLALGKRIHPELPEIEYVDVATAPMAISQMDAGGFDLAILDGEATPAGG MGVAKQVKDEIDDCPPILVLTGRADDAWLAKWSRAEAAVPHPIDPIRLSDAVVSLLRAPAQ*
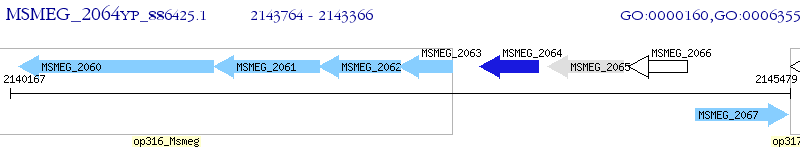
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2064 | - | - | 100% (132) | two-component system response regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3167 | - | 9e-53 | 74.22% (128) | response regulator |
| M. gilvum PYR-GCK | Mflv_4480 | - | 3e-60 | 84.80% (125) | response regulator receiver protein |
| M. tuberculosis H37Rv | Rv3143 | - | 9e-53 | 74.22% (128) | response regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2133 | - | 1e-36 | 58.14% (129) | response regulator |
| M. marinum M | MMAR_1486 | - | 3e-52 | 73.44% (128) | two-component system response regulator |
| M. avium 104 | MAV_4032 | - | 1e-50 | 68.99% (129) | two-component system response regulator |
| M. thermoresistible (build 8) | TH_0638 | - | 1e-50 | 72.80% (125) | PROBABLE RESPONSE REGULATOR |
| M. ulcerans Agy99 | MUL_2453 | - | 2e-52 | 73.44% (128) | two-component system response regulator |
| M. vanbaalenii PYR-1 | Mvan_1886 | - | 3e-60 | 84.92% (126) | response regulator receiver protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3167|M.bovis_AF2122/97 ----------------MPDS---STALRILVYSDNVQTRERVMRALGKRL
Rv3143|M.tuberculosis_H37Rv ----------------MPDS---STALRILVYSDNVQTRERVMRALGKRL
MMAR_1486|M.marinum_M MVPPGAGQPETSSICTVADS---SEDLRILVYSDNVQTREQVVRALGKRL
MUL_2453|M.ulcerans_Agy99 ----------------MADS---SEDLRILVYSDNVQTREQVVRALGKRL
MAV_4032|M.avium_104 ------------MSDSTPQS---SVALRILVYSSNAQTRERVMRALGRHL
Mflv_4480|M.gilvum_PYR-GCK ------------MVVPSAET---RGVLRVLVYSDNPRTREQVRLALGRRI
Mvan_1886|M.vanbaalenii_PYR-1 ------------MAAPSSESSRAQDPLRILVYSDNPRTREQVRLALGRRI
MSMEG_2064|M.smegmatis_MC2_155 ------------MAG--------SAPLRILVYSDNPRTREQVRLALGKRI
TH_0638|M.thermoresistible__bu ------------VSRPVDD----AAALRILVYSDDPRTRDAVRTALGTRL
MAB_2133|M.abscessus_ATCC_1997 ------------------------MALQVLVYSDDITVRRKVIQGIGARP
*::****.: .* * .:* :
Mb3167|M.bovis_AF2122/97 HPDLPDLTYVEVATGPMVIRQMDRG----GIDLAILDGEATPTGGMGIAK
Rv3143|M.tuberculosis_H37Rv HPDLPDLTYVEVATGPMVIRQMDRG----GIDLAILDGEATPTGGMGIAK
MMAR_1486|M.marinum_M HPDLPALTYVEVATGPMVLRRMDEG----GIDLAILDGEATPTGGMGIAK
MUL_2453|M.ulcerans_Agy99 HPDLPALTYVEVATGPMVLRRMDEG----GIDLAILDGEATPTGGMGIAK
MAV_4032|M.avium_104 HPDLPELSYVEVATGPMVVKTMDQG----GIDLAILDGEATPTGGMGIAK
Mflv_4480|M.gilvum_PYR-GCK HPELPELDYVEVATGPMVVRQMDAG----GFDLAILDGEATPVGGMGIAK
Mvan_1886|M.vanbaalenii_PYR-1 HPELPELSYLEVATGPMVIRQMDEG----GFDLAILDGEAAPVGGMGIAK
MSMEG_2064|M.smegmatis_MC2_155 HPELPEIEYVDVATAPMAISQMDAG----GFDLAILDGEATPAGGMGVAK
TH_0638|M.thermoresistible__bu HPDLPELGYQEVATEPMVHEHLRSG----HFDLVILDGEATPAGGMGIAK
MAB_2133|M.abscessus_ATCC_1997 DSALPPLEFVEVATGPMVIERLAAPGSLPGIDLAILDGEATPFGGMGVAK
.. ** : : :*** **. : :**.******:* ****:**
Mb3167|M.bovis_AF2122/97 QLKDELASCPPILVLTGRPDDTWLASWSRAEAAVPHPVDPIVLGRTVLSL
Rv3143|M.tuberculosis_H37Rv QLKDELASCPPILVLTGRPDDTWLASWSRAEAAVPHPVDPIVLGRTVLSL
MMAR_1486|M.marinum_M QLKDELDNCPPLVVLTGRPDDAWLASWSRAEAAVPHPIDPIVLGRTVLGL
MUL_2453|M.ulcerans_Agy99 QLKDELDNCPPLVVLTGRPDDAWLASWSRAEAAVPHPIDPIVLGRTVLGL
MAV_4032|M.avium_104 QLKDELDACPPLVVLTGRPDDAWLASWSRAEAAVPHPIDPILLGRTVLNL
Mflv_4480|M.gilvum_PYR-GCK QLKDELDVCPPILVLTGRPDDAWLARWSRAEAAVPHPIDPIRLGDAVVSL
Mvan_1886|M.vanbaalenii_PYR-1 QLKDEIDDCPPVLVLTRRADDAWLARWSRAEAAVPHPIDPIRLGDAVVSL
MSMEG_2064|M.smegmatis_MC2_155 QVKDEIDDCPPILVLTGRADDAWLAKWSRAEAAVPHPIDPIRLSDAVVSL
TH_0638|M.thermoresistible__bu QLKDELADCPPVLVLTGRRDDAWLARWSRAEAAVPQPIDPIELGDAVLSL
MAB_2133|M.abscessus_ATCC_1997 QVKDEVDAPPPIVILTGRPQDNWLASWSRAEAVVPRPIDPMRLTATVIDL
*:***: **:::** * :* *** ******.**:*:**: * :*:.*
Mb3167|M.bovis_AF2122/97 LRAPAH---
Rv3143|M.tuberculosis_H37Rv LRAPAH---
MMAR_1486|M.marinum_M LRAPAL---
MUL_2453|M.ulcerans_Agy99 LRAPAL---
MAV_4032|M.avium_104 LRTPVR---
Mflv_4480|M.gilvum_PYR-GCK LRPAA----
Mvan_1886|M.vanbaalenii_PYR-1 LRPSA----
MSMEG_2064|M.smegmatis_MC2_155 LRAPAQ---
TH_0638|M.thermoresistible__bu LRTRTS---
MAB_2133|M.abscessus_ATCC_1997 LGSAAASRR
* . .