For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEVFLELGQRPDEAGPPISGPATYPDDVTESLRADAEQIIARYPDARSALLPLLHLVQAQDGYLTPAGIG FCAAQLGLTEAEVTAVATFYSMYRRTPTGDYLVGVCTNTLCAIMGGDAILEALEDHLGVHPGQTTPDGRV TLEHVECNAACDYAPVVMVNWEFYDNQTPSSARDLVDGLRSGSPPPPTRGSLCTFRETARTLAGLTDPNA PGGAPGAATLAGLRLARERGMTAPTPPQTNGSAS*
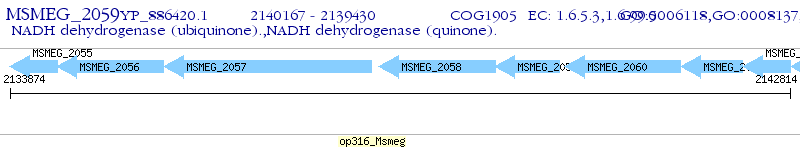
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2059 | - | - | 100% (245) | NADH dehydrogenase subunit E |
| M. smegmatis MC2 155 | MSMEG_0159 | - | 3e-17 | 33.57% (140) | formate dehydrogenase, gamma subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3173 | nuoE | 1e-87 | 65.83% (240) | NADH dehydrogenase subunit E |
| M. gilvum PYR-GCK | Mflv_4485 | - | 1e-103 | 78.63% (234) | NADH dehydrogenase subunit E |
| M. tuberculosis H37Rv | Rv3149 | nuoE | 1e-87 | 65.83% (240) | NADH dehydrogenase subunit E |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2138 | - | 2e-82 | 65.67% (233) | NADH-quinone oxidoreductase, E subunit NuoE |
| M. marinum M | MMAR_1479 | nuoE | 3e-85 | 65.83% (240) | NADH dehydrogenase I (chain E) NuoE (NADH- ubiquinone |
| M. avium 104 | MAV_4037 | - | 1e-91 | 67.63% (241) | NADH dehydrogenase subunit E |
| M. thermoresistible (build 8) | TH_0633 | nuoE | 1e-101 | 75.82% (244) | PROBABLE NADH DEHYDROGENASE I (CHAIN E) NUOE |
| M. ulcerans Agy99 | MUL_2463 | nuoE | 2e-86 | 66.25% (240) | NADH dehydrogenase subunit E |
| M. vanbaalenii PYR-1 | Mvan_1881 | - | 1e-104 | 75.81% (248) | NADH dehydrogenase subunit E |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3173|M.bovis_AF2122/97 MTQPPGQPVFIR---LGPPPDEPNQFVVEG---------APRSYPPDVLA
Rv3149|M.tuberculosis_H37Rv MTQPPGQPVFIR---LGPPPDEPNQFVVEG---------APRSYPPDVLA
MMAR_1479|M.marinum_M MNAPQSQPVFVR---LGPPPEEPNQFTVEG---------APQAYSPDALA
MUL_2463|M.ulcerans_Agy99 MNAPQSQPVFVR---LGPPPEEPNQFTVEG---------APQAYPPDALA
MAV_4037|M.avium_104 MTGPQGQPVFLR---LGPPPDEPNAFVVEG---------APTSYPPEVRA
Mflv_4485|M.gilvum_PYR-GCK --------MFL---ELGQRPDEPG-PPIHG----------PATYPAAVHE
Mvan_1881|M.vanbaalenii_PYR-1 ------MSVFL---ELGQRPDEPG-PPIHG----------PTAYPAEVTE
MSMEG_2059|M.smegmatis_MC2_155 -----MSEVFL---ELGQRPDEAG-PPISG----------PATYPDDVTE
TH_0633|M.thermoresistible__bu MTGGNGTPVFISVAELGQRPDEDG-PPIAGPASYGPPIAGPASYPPEVTE
MAB_2138|M.abscessus_ATCC_1997 -----MTEVFVE---LGSRPSEDE-GRFAG----------RKHYPAEVVS
:*: ** *.* . * *. .
Mb3173|M.bovis_AF2122/97 RLEVDAKEIIGRYPDRRSALLPLLHLVQGEDSYLTPAGLRFCADQLGLTG
Rv3149|M.tuberculosis_H37Rv RLEVDAKEIIGRYPDRRSALLPLLHLVQGEDSYLTPAGLRFCADQLGLTG
MMAR_1479|M.marinum_M RLEIEAKEIIGRYPDQRSALLPLLHLVQGEDSYLTPAGLQFCANQLGLTG
MUL_2463|M.ulcerans_Agy99 RLEIEAKEIIGRYPDQRSALLPLLHLVQGEDSYLTPAGLQFCANQLGLTG
MAV_4037|M.avium_104 RLEVDAKEIMGRYPDKRSALLPLLHLVQSQDSYLTPAGLEFCGEQLGLTG
Mflv_4485|M.gilvum_PYR-GCK RLTADAATIIARYPQTRSALLPLLHLVQAQDGCLTPAGIAFCADRLGLTD
Mvan_1881|M.vanbaalenii_PYR-1 RLAADAATIIGRYPQSRSALLPLLHLVQAEDGYLTPAGIAFCAGQLGLTH
MSMEG_2059|M.smegmatis_MC2_155 SLRADAEQIIARYPDARSALLPLLHLVQAQDGYLTPAGIGFCAAQLGLTE
TH_0633|M.thermoresistible__bu RLAADAATIIARYPQARSALLPLLHLVQGEDGYLTRAGIAFCADQLSLSA
MAB_2138|M.abscessus_ATCC_1997 RLAIDAKEILDRYPSRRSALLPLLHLVQSEDGYVTTAGIEFCARQVELTS
* :* *: ***. ************.:*. :* **: **. :: *:
Mb3173|M.bovis_AF2122/97 AEVSAVASFYTMYRRRPTGEYLVGVCTNTLCAVMGGDAIFDRLKEHLGVG
Rv3149|M.tuberculosis_H37Rv AEVSAVASFYTMYRRRPTGEYLVGVCTNTLCAVMGGDAIFDRLKEHLGVG
MMAR_1479|M.marinum_M AEVSAVASFYTMYRRGPTGEYLVGVCTNTLCAVMGGDAIFETLKDHLGVG
MUL_2463|M.ulcerans_Agy99 AEVSAVASFYTMYRRGPTGEYLVGVCTNTLCAVMGGDAIFETLKDHLGVG
MAV_4037|M.avium_104 AEVSAVASFYTMYRRGPTGDYLVGVCTNTLCAIMGGDAIFDALKEHLGIG
Mflv_4485|M.gilvum_PYR-GCK AEVTAVATFYSMYRRTPTGEYLVGVCTNTLCAVMGGDAILESLEQHLDIA
Mvan_1881|M.vanbaalenii_PYR-1 AEVTAVATFYSMYRREPTGEYLVGVCTNTLCAVMGGDAILESLESHLDIA
MSMEG_2059|M.smegmatis_MC2_155 AEVTAVATFYSMYRRTPTGDYLVGVCTNTLCAIMGGDAILEALEDHLGVH
TH_0633|M.thermoresistible__bu AEVAAVATFYSMYRRRPTGDYLVGVCTNTLCAIMGGDAILDALQEHLGVP
MAB_2138|M.abscessus_ATCC_1997 AEVTAVASFYSMYRREPTGDYLVGVCTNTLCAVLGGDAILARLAGELGVR
***:***:**:**** ***:************::*****: * .*.:
Mb3173|M.bovis_AF2122/97 HDETTSDGVVTLQHIECNAACDYAPVVMVNWEFFDNQTPESARELVDSLR
Rv3149|M.tuberculosis_H37Rv HDETTSDGVVTLQHIECNAACDYAPVVMVNWEFFDNQTPESARELVDSLR
MMAR_1479|M.marinum_M NDETTSDGSVTLGHIECNAACDYAPVVMVNWEFFDNQTPESARELVDSLR
MUL_2463|M.ulcerans_Agy99 NDETTSDGSVTLGHIECNAACDYAPVVMVNWEFFDNQTPESARELVDSLR
MAV_4037|M.avium_104 NDETTPDGSVTLQHIECNAACDYAPVVMVNWEFFDNQTPESARELVDSLR
Mflv_4485|M.gilvum_PYR-GCK PGQTTADGRITLEHVECNAACDFAPVVMVNWDFFDNQTPASARELVDGLR
Mvan_1881|M.vanbaalenii_PYR-1 PGQTTADGTITLEHVECNAACDYAPVVMVNWDFFDNQTPSSVRDLVDSLR
MSMEG_2059|M.smegmatis_MC2_155 PGQTTPDGRVTLEHVECNAACDYAPVVMVNWEFYDNQTPSSARDLVDGLR
TH_0633|M.thermoresistible__bu AGGTTEDGAVTLEHIECNAACDYAPVVMVNWEFFDNQTPASARELVDGLR
MAB_2138|M.abscessus_ATCC_1997 PGGTTEDGKVTLEHIECNAGCDYAPVLMVNWEFFDNQTPDGAAALVQGLR
. ** ** :** *:****.**:***:****:*:***** .. **:.**
Mb3173|M.bovis_AF2122/97 SDTPKAPTRGAPLCGFRQTSRILAGLPDQRPDEGQGG-PGAPTLAGLQVA
Rv3149|M.tuberculosis_H37Rv SDTPKAPTRGAPLCGFRQTSRILAGLPDQRPDEGQGG-PGAPTLAGLQVA
MMAR_1479|M.marinum_M SGEPIAPTRGAPLCGFRQTSRILAGLPDTRPDQGQGG-AGAATLAGLRVA
MUL_2463|M.ulcerans_Agy99 SGEPIAPTRGAPLCGFRQTSRILAGLPDTRPDQGQGG-AGAATLAGLRVA
MAV_4037|M.avium_104 AGKPRHPTRGAPLCVFRETSRILAGLPDERPDQGQGG-AGAATLAGLKVA
Mflv_4485|M.gilvum_PYR-GCK SGQTPAPSRGGSLCTFRETSRILAGLDGP-DSATAGT-VGEATLAGLRFA
Mvan_1881|M.vanbaalenii_PYR-1 SGEKPTPSRSGALCTFRETARILAGLSNPGDAPDAGR-VGAATLAGLRVA
MSMEG_2059|M.smegmatis_MC2_155 SGSPPPPTR-GSLCTFRETARTLAGLTDP--NAPGGA-PGAATLAGLRLA
TH_0633|M.thermoresistible__bu SGRPPEPTRGAPLCSFRQTARILAGLPDP-RPKTPGG-PGEATLAGLRVA
MAB_2138|M.abscessus_ATCC_1997 RGHVPAPRRGAPPCTFKETARILAGFADERPEAVAAAQPGDSTLAGLRLA
. * * .. * *::*:* ***: . . * .*****:.*
Mb3173|M.bovis_AF2122/97 RKNDMQAP--PTPGADE---------------------------------
Rv3149|M.tuberculosis_H37Rv RKNDMQAP--PTPGADE---------------------------------
MMAR_1479|M.marinum_M QDNDMQAP--PAPGGQ----------------------------------
MUL_2463|M.ulcerans_Agy99 QDNDMQAP--PAPGGQ----------------------------------
MAV_4037|M.avium_104 KEHGMQAP--YPPEGQQ---------------------------------
Mflv_4485|M.gilvum_PYR-GCK REHRMAAPAAGSPSDTTAGGGGQEQAVAAESVKNEPAPAPEADVPARSGT
Mvan_1881|M.vanbaalenii_PYR-1 QERAMAAPDHGAPPDT----------------------------------
MSMEG_2059|M.smegmatis_MC2_155 RERGMTAP---TPPQT----------------------------------
TH_0633|M.thermoresistible__bu RERGMSAP---EPTS-----------------------------------
MAB_2138|M.abscessus_ATCC_1997 QDVQRNVQYKADGMRP----------------------------------
:. .
Mb3173|M.bovis_AF2122/97 -----------
Rv3149|M.tuberculosis_H37Rv -----------
MMAR_1479|M.marinum_M -----------
MUL_2463|M.ulcerans_Agy99 -----------
MAV_4037|M.avium_104 -----------
Mflv_4485|M.gilvum_PYR-GCK AETDPEATDAP
Mvan_1881|M.vanbaalenii_PYR-1 ---DPKAADDR
MSMEG_2059|M.smegmatis_MC2_155 ---NGSAS---
TH_0633|M.thermoresistible__bu -----------
MAB_2138|M.abscessus_ATCC_1997 -----------