For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKRTTTGETSVATLVREIAADHRDHRGPLLPILHAVQERLGCVPAEAVPVLAEELNLSRADVHGVITFYH DFRSEPAGRTTVRVCRAEACQALGASRLVAHLQDRHGVQLGDATDDGTLTAEQVFCLGNCALGPSAQVDG RLYGRLDEARLSALIDSAVTR
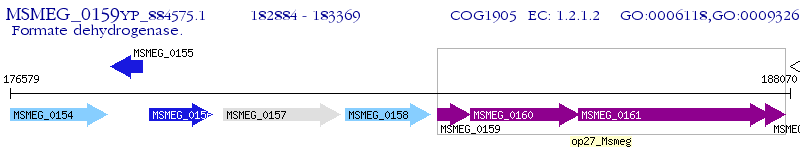
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0159 | - | - | 100% (161) | formate dehydrogenase, gamma subunit |
| M. smegmatis MC2 155 | MSMEG_2059 | - | 2e-17 | 33.57% (140) | NADH dehydrogenase subunit E |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3173 | nuoE | 4e-15 | 28.87% (142) | NADH dehydrogenase subunit E |
| M. gilvum PYR-GCK | Mflv_4485 | - | 1e-16 | 31.65% (139) | NADH dehydrogenase subunit E |
| M. tuberculosis H37Rv | Rv3149 | nuoE | 4e-15 | 28.87% (142) | NADH dehydrogenase subunit E |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2138 | - | 3e-17 | 32.14% (140) | NADH-quinone oxidoreductase, E subunit NuoE |
| M. marinum M | MMAR_1479 | nuoE | 1e-14 | 29.58% (142) | NADH dehydrogenase I (chain E) NuoE (NADH- ubiquinone |
| M. avium 104 | MAV_4037 | - | 2e-14 | 28.17% (142) | NADH dehydrogenase subunit E |
| M. thermoresistible (build 8) | TH_0633 | nuoE | 1e-17 | 33.09% (139) | PROBABLE NADH DEHYDROGENASE I (CHAIN E) NUOE |
| M. ulcerans Agy99 | MUL_2463 | nuoE | 8e-15 | 29.58% (142) | NADH dehydrogenase subunit E |
| M. vanbaalenii PYR-1 | Mvan_1881 | - | 2e-16 | 30.72% (153) | NADH dehydrogenase subunit E |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3173|M.bovis_AF2122/97 MTQPPGQPVFIR---LGPPPDEPNQFVVEGA--------PRSYPPDVLAR
Rv3149|M.tuberculosis_H37Rv MTQPPGQPVFIR---LGPPPDEPNQFVVEGA--------PRSYPPDVLAR
MMAR_1479|M.marinum_M MNAPQSQPVFVR---LGPPPEEPNQFTVEGA--------PQAYSPDALAR
MUL_2463|M.ulcerans_Agy99 MNAPQSQPVFVR---LGPPPEEPNQFTVEGA--------PQAYPPDALAR
MAV_4037|M.avium_104 MTGPQGQPVFLR---LGPPPDEPNAFVVEGA--------PTSYPPEVRAR
TH_0633|M.thermoresistible__bu MTGGNGTPVFISVAELGQRPDEDGPPIAGPASYGPPIAGPASYPPEVTER
Mflv_4485|M.gilvum_PYR-GCK --------MFLE---LGQRPDEPGPPIHGPA----------TYPAAVHER
Mvan_1881|M.vanbaalenii_PYR-1 ------MSVFLE---LGQRPDEPGPPIHGPT----------AYPAEVTER
MAB_2138|M.abscessus_ATCC_1997 -----MTEVFVE---LGSRPSEDEGRFAGRK----------HYPAEVVSR
MSMEG_0159|M.smegmatis_MC2_155 ----------------------------------------MKRTTTGETS
..
Mb3173|M.bovis_AF2122/97 LEVDAKEIIGRYPDRRSALLPLLHLVQGEDSYLTPAGLRFCADQLGLTGA
Rv3149|M.tuberculosis_H37Rv LEVDAKEIIGRYPDRRSALLPLLHLVQGEDSYLTPAGLRFCADQLGLTGA
MMAR_1479|M.marinum_M LEIEAKEIIGRYPDQRSALLPLLHLVQGEDSYLTPAGLQFCANQLGLTGA
MUL_2463|M.ulcerans_Agy99 LEIEAKEIIGRYPDQRSALLPLLHLVQGEDSYLTPAGLQFCANQLGLTGA
MAV_4037|M.avium_104 LEVDAKEIMGRYPDKRSALLPLLHLVQSQDSYLTPAGLEFCGEQLGLTGA
TH_0633|M.thermoresistible__bu LAADAATIIARYPQARSALLPLLHLVQGEDGYLTRAGIAFCADQLSLSAA
Mflv_4485|M.gilvum_PYR-GCK LTADAATIIARYPQTRSALLPLLHLVQAQDGCLTPAGIAFCADRLGLTDA
Mvan_1881|M.vanbaalenii_PYR-1 LAADAATIIGRYPQSRSALLPLLHLVQAEDGYLTPAGIAFCAGQLGLTHA
MAB_2138|M.abscessus_ATCC_1997 LAIDAKEILDRYPSRRSALLPLLHLVQSEDGYVTTAGIEFCARQVELTSA
MSMEG_0159|M.smegmatis_MC2_155 VATLVREIAADHRDHRGPLLPILHAVQERLGCVPAEAVPVLAEELNLSRA
: . * : . *..***:** ** . . :. .: . . .: *: *
Mb3173|M.bovis_AF2122/97 EVSAVASFYTMYRRRPTGEYLVGVCTNTLCAVMGGDAIFDRLKEHLGVGH
Rv3149|M.tuberculosis_H37Rv EVSAVASFYTMYRRRPTGEYLVGVCTNTLCAVMGGDAIFDRLKEHLGVGH
MMAR_1479|M.marinum_M EVSAVASFYTMYRRGPTGEYLVGVCTNTLCAVMGGDAIFETLKDHLGVGN
MUL_2463|M.ulcerans_Agy99 EVSAVASFYTMYRRGPTGEYLVGVCTNTLCAVMGGDAIFETLKDHLGVGN
MAV_4037|M.avium_104 EVSAVASFYTMYRRGPTGDYLVGVCTNTLCAIMGGDAIFDALKEHLGIGN
TH_0633|M.thermoresistible__bu EVAAVATFYSMYRRRPTGDYLVGVCTNTLCAIMGGDAILDALQEHLGVPA
Mflv_4485|M.gilvum_PYR-GCK EVTAVATFYSMYRRTPTGEYLVGVCTNTLCAVMGGDAILESLEQHLDIAP
Mvan_1881|M.vanbaalenii_PYR-1 EVTAVATFYSMYRREPTGEYLVGVCTNTLCAVMGGDAILESLESHLDIAP
MAB_2138|M.abscessus_ATCC_1997 EVTAVASFYSMYRREPTGDYLVGVCTNTLCAVLGGDAILARLAGELGVRP
MSMEG_0159|M.smegmatis_MC2_155 DVHGVITFYHDFRSEPAGRTTVRVCRAEACQALGASRLVAHLQDRHGVQL
:* .* :** :* *:* * ** * :*.. :. * . .:
Mb3173|M.bovis_AF2122/97 DETTSDGVVTLQHIECNAACDYAPVVMVNWEFFDNQTPESARELVDSLRS
Rv3149|M.tuberculosis_H37Rv DETTSDGVVTLQHIECNAACDYAPVVMVNWEFFDNQTPESARELVDSLRS
MMAR_1479|M.marinum_M DETTSDGSVTLGHIECNAACDYAPVVMVNWEFFDNQTPESARELVDSLRS
MUL_2463|M.ulcerans_Agy99 DETTSDGSVTLGHIECNAACDYAPVVMVNWEFFDNQTPESARELVDSLRS
MAV_4037|M.avium_104 DETTPDGSVTLQHIECNAACDYAPVVMVNWEFFDNQTPESARELVDSLRA
TH_0633|M.thermoresistible__bu GGTTEDGAVTLEHIECNAACDYAPVVMVNWEFFDNQTPASARELVDGLRS
Mflv_4485|M.gilvum_PYR-GCK GQTTADGRITLEHVECNAACDFAPVVMVNWDFFDNQTPASARELVDGLRS
Mvan_1881|M.vanbaalenii_PYR-1 GQTTADGTITLEHVECNAACDYAPVVMVNWDFFDNQTPSSVRDLVDSLRS
MAB_2138|M.abscessus_ATCC_1997 GGTTEDGKVTLEHIECNAGCDYAPVLMVNWEFFDNQTPDGAAALVQGLRR
MSMEG_0159|M.smegmatis_MC2_155 GDATDDGTLTAEQVFCLGNCALGPSAQVDGRLYGRLDEARLSALIDSAVT
. :* ** :* :: * . * .* *: ::.. *::.
Mb3173|M.bovis_AF2122/97 DTPKAPTRGAPLCGFRQTSRILAGLPDQRPDEGQGG-PGAPTLAGLQVAR
Rv3149|M.tuberculosis_H37Rv DTPKAPTRGAPLCGFRQTSRILAGLPDQRPDEGQGG-PGAPTLAGLQVAR
MMAR_1479|M.marinum_M GEPIAPTRGAPLCGFRQTSRILAGLPDTRPDQGQGG-AGAATLAGLRVAQ
MUL_2463|M.ulcerans_Agy99 GEPIAPTRGAPLCGFRQTSRILAGLPDTRPDQGQGG-AGAATLAGLRVAQ
MAV_4037|M.avium_104 GKPRHPTRGAPLCVFRETSRILAGLPDERPDQGQGG-AGAATLAGLKVAK
TH_0633|M.thermoresistible__bu GRPPEPTRGAPLCSFRQTARILAGLPDPRP-KTPGG-PGEATLAGLRVAR
Mflv_4485|M.gilvum_PYR-GCK GQTPAPSRGGSLCTFRETSRILAGLDGP-DSATAGT-VGEATLAGLRFAR
Mvan_1881|M.vanbaalenii_PYR-1 GEKPTPSRSGALCTFRETARILAGLSNPGDAPDAGR-VGAATLAGLRVAQ
MAB_2138|M.abscessus_ATCC_1997 GHVPAPRRGAPPCTFKETARILAGFADERPEAVAAAQPGDSTLAGLRLAQ
MSMEG_0159|M.smegmatis_MC2_155 R-------------------------------------------------
Mb3173|M.bovis_AF2122/97 KNDMQAPPTPGADE------------------------------------
Rv3149|M.tuberculosis_H37Rv KNDMQAPPTPGADE------------------------------------
MMAR_1479|M.marinum_M DNDMQAPPAPGGQ-------------------------------------
MUL_2463|M.ulcerans_Agy99 DNDMQAPPAPGGQ-------------------------------------
MAV_4037|M.avium_104 EHGMQAPYPPEGQQ------------------------------------
TH_0633|M.thermoresistible__bu ERGMSAPEPTS---------------------------------------
Mflv_4485|M.gilvum_PYR-GCK EHRMAAPAAGSPSDTTAGGGGQEQAVAAESVKNEPAPAPEADVPARSGTA
Mvan_1881|M.vanbaalenii_PYR-1 ERAMAAPDHGAPPDT-----------------------------------
MAB_2138|M.abscessus_ATCC_1997 DVQRNVQYKADGMRP-----------------------------------
MSMEG_0159|M.smegmatis_MC2_155 --------------------------------------------------
Mb3173|M.bovis_AF2122/97 ----------
Rv3149|M.tuberculosis_H37Rv ----------
MMAR_1479|M.marinum_M ----------
MUL_2463|M.ulcerans_Agy99 ----------
MAV_4037|M.avium_104 ----------
TH_0633|M.thermoresistible__bu ----------
Mflv_4485|M.gilvum_PYR-GCK ETDPEATDAP
Mvan_1881|M.vanbaalenii_PYR-1 --DPKAADDR
MAB_2138|M.abscessus_ATCC_1997 ----------
MSMEG_0159|M.smegmatis_MC2_155 ----------