For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGSDTTAYSTARTVEDFTRNLQAVNTRIGAALQRAGREPGEVRLLPVSKTVDEDRLRVAAAAGCTMFGE NKVQEAMRKWRNLSDLDVEWSVIGHLQTNKAKDVAEFAAEFQALDNPRAAAALDRRLQALGRQLDVYVQV NTSGEESKYGLAPDDVIPFLKTLPAYTALRVRGLMTIAVFSTDPERVRPCFRLLRSLRDQARDLDLIGPG ELSMGMSGDYELAIAEGSTCVRVGQAIFGARPTPDSQYWPGN
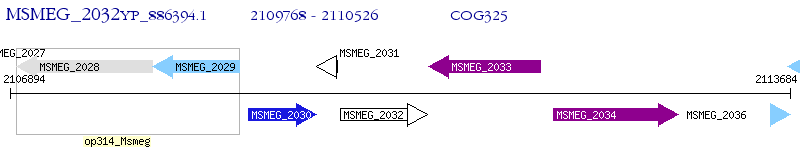
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2032 | - | - | 100% (252) | hypothetical protein MSMEG_2032 |
| M. smegmatis MC2 155 | MSMEG_4220 | - | 3e-29 | 35.14% (259) | hypothetical protein MSMEG_4220 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2172c | - | 1e-28 | 33.85% (257) | hypothetical protein Mb2172c |
| M. gilvum PYR-GCK | Mflv_2993 | - | 2e-27 | 33.85% (257) | alanine racemase domain-containing protein |
| M. tuberculosis H37Rv | Rv2148c | - | 1e-28 | 33.85% (257) | hypothetical protein Rv2148c |
| M. leprae Br4923 | MLBr_00919 | - | 6e-29 | 35.29% (238) | putative DNA-binding protein |
| M. abscessus ATCC 19977 | MAB_2011 | - | 1e-31 | 36.60% (235) | hypothetical protein MAB_2011 |
| M. marinum M | MMAR_3188 | - | 9e-27 | 32.65% (245) | hypothetical protein MMAR_3188 |
| M. avium 104 | MAV_2342 | - | 1e-29 | 33.84% (263) | hypothetical protein MAV_2342 |
| M. thermoresistible (build 8) | TH_2376 | - | 6e-24 | 34.24% (257) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3496 | - | 1e-25 | 31.15% (244) | hypothetical protein MUL_3496 |
| M. vanbaalenii PYR-1 | Mvan_3518 | - | 9e-30 | 35.22% (247) | alanine racemase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3188|M.marinum_M MTAMVVDISAQGERQSELTHALAVVRSRLAAAAEAAGRKVSEIELLPITK
MUL_3496|M.ulcerans_Agy99 ---MVVDISAQGERQSELTHALALVRSRLAAAAEAAGRKVSEIELLPITK
Mb2172c|M.bovis_AF2122/97 ---MAADLSAYPDRESELTHALAAMRSRLAAAAEAAGRNVGEIELLPITK
Rv2148c|M.tuberculosis_H37Rv ---MAADLSAYPDRESELTHALAAMRSRLAAAAEAAGRNVGEIELLPITK
MLBr_00919|M.leprae_Br4923 MTAMAADIDLQGDRESELMHALATVRSRLAAASQAAGRNVGEIELLPISK
MAV_2342|M.avium_104 MTAGTPAPDSAGDRETELTHALAAVRSRLAAAAEAAGRNVAEIEVLPVTK
Mflv_2993|M.gilvum_PYR-GCK ---------MTSEREAELRAALDALRVRLARAAESAGRDVAEIELLPVIK
Mvan_3518|M.vanbaalenii_PYR-1 ---------MTAARDAELSLALTALRLRVARAAEAAGRDAGDIELLPVTK
TH_2376|M.thermoresistible__bu -----------------LAVALAALRARLEQAAVAAGRAADEIELLPITK
MAB_2011|M.abscessus_ATCC_1997 -----------MTRAEELSAALCSVRERVRRAAEIAGRDPGEIQLLPVTK
MSMEG_2032|M.smegmatis_MC2_155 MTGSDTTAYSTARTVEDFTRNLQAVNTRIGAALQRAGREPGEVRLLPVSK
: * :. *: * *** ::.:**: *
MMAR_3188|M.marinum_M FFPATDVAILARLGCRAVGESRDQEASAKVAELARLFSAPIRSGSRGIQW
MUL_3496|M.ulcerans_Agy99 FFPATDVAILARLGCRAVGESRDQEASAKVAELARMFSAPMRSGSRGIQW
Mb2172c|M.bovis_AF2122/97 FFPATDVAILFRLGCRSVGESREQEASAKMAELNRLLAAAELGHSGGVHW
Rv2148c|M.tuberculosis_H37Rv FFPATDVAILFRLGCRSVGESREQEASAKMAELNRLLAAAELGHSGGVHW
MLBr_00919|M.leprae_Br4923 FFPATDVAILSRLGCRSVGESRAQEASTKAAEFAELLGVSREEKS-SIHW
MAV_2342|M.avium_104 FFPATDVVTLSRLGCRAVGESRDQEATAKVAEVTRLLATSARTNVAIPRW
Mflv_2993|M.gilvum_PYR-GCK FFPASDVLALYRLGCDAFAESRDQEATAKIEDVAR-VVGSR--EDTPIRW
Mvan_3518|M.vanbaalenii_PYR-1 FFPASDVVALHRLGCSAFGESRDQEAAAKVEEVGG-LVG-----NDPIRW
TH_2376|M.thermoresistible__bu FFPASDVLILHGLGISAFGESREQEASAKIAEVNRNLSVEQ--NASPIRW
MAB_2011|M.abscessus_ATCC_1997 FFPASDVEELYRLGCREFGESRDQEAAAKVALLDQ-------AGHADIRW
MSMEG_2032|M.smegmatis_MC2_155 TVDEDRLRVAAAAGCTMFGENKVQEAMRKWRNLSD----------LDVEW
. : * ..*.: *** * . .*
MMAR_3188|M.marinum_M HMVGQIQRNKVRSLANWAHTAHSIDTPQLVTALDRAITTAIADRRRGNPL
MUL_3496|M.ulcerans_Agy99 HMVGQIQRNKVRSLANWAHTAHSIDTPQLVTALDRAITTAIADRRRGNPL
Mb2172c|M.bovis_AF2122/97 HMVGRIQRNKAGSLARWAHTAHSVDSSRLVTALDRAVVAALAEHRRGERL
Rv2148c|M.tuberculosis_H37Rv HMVGRIQRNKAGSLARWAHTAHSVDSSRLVTALDRAVVAALAEHRRGERL
MLBr_00919|M.leprae_Br4923 HMVGQIQRNKVRSLAQWAHTAHSIDSLQLVAALDRAVAAALAGGRREQPL
MAV_2342|M.avium_104 HMVGHIQRNKARSVARWAHTAHSVDSTHLVSALDRGVAAALADGGRIDPM
Mflv_2993|M.gilvum_PYR-GCK HMVGRIQRNKARSVAEWAYAAHSVDSVKVVAALERSASTALEEGRRTEPL
Mvan_3518|M.vanbaalenii_PYR-1 HMVGRIQRNKARSVAQWAYAAHSVDSPKVIAALDRAAEEALDDGRRPAPL
TH_2376|M.thermoresistible__bu HMVGRIQRNKARAVARWAYAAHSVDNAKLVAALSRGAVQALDEGVRTAPV
MAB_2011|M.abscessus_ATCC_1997 HMVGNLQRNKAKSVAHWAHSVHSVDNARLVTALDG----ARGAGEKREPL
MSMEG_2032|M.smegmatis_MC2_155 SVIGHLQTNKAKDVAEFAAEFQALDNPRAAAALDR------RLQALGRQL
::*.:* **. :*.:* :::*. : :**. :
MMAR_3188|M.marinum_M RVYVQVSLDGDVSRGGVDMTDPGAVDRICAQVSDAQSLELIGLMGVPPRD
MUL_3496|M.ulcerans_Agy99 RVHVQVSLDGDVSRGGVDMTDPGAVDRICAQVSDSQSLELIGLMGVPPRD
Mb2172c|M.bovis_AF2122/97 RVYVQVSLDGDGSRGGVDSTTPGAVDRICAQVQESEGLELVGLMGIPPLD
Rv2148c|M.tuberculosis_H37Rv RVYVQVSLDGDGSRGGVDSTTPGAVDRICAQVQESEGLELVGLMGIPPLD
MLBr_00919|M.leprae_Br4923 QVYVQISLDGDISRGGVNVTAPGAVDRVCAQVEESKSLELVGLMGIPPLG
MAV_2342|M.avium_104 RVYVQVSLDGDESRGGVDISRPGAVDEVCAQVAAANNLDLVGLMAIPPLN
Mflv_2993|M.gilvum_PYR-GCK RVFAQLSLDGDEARGGVDVDATERVDELCAAMDRAGGLTFAGMMAIPPLG
Mvan_3518|M.vanbaalenii_PYR-1 RVYLQVSLDGDEARGGVDVAATDRIDDLCGAVDSARGLEFVGLMAIPPLN
TH_2376|M.thermoresistible__bu RVYVQVSLDGDERRGGVDVGRPQLVDELCAAVHDAAGLELVGLMGIPPLN
MAB_2011|M.abscessus_ATCC_1997 GIYLQVSLDGDNARGGVDIAAADEIDQLCAQVVASEHLELAGLMALPPRE
MSMEG_2032|M.smegmatis_MC2_155 DVYVQVNTSGEESKYGL---APDDVIPFLKTLPAYTALRVRGLMTIAVFS
:. *:. .*: : *: . : . : * . *:* :.
MMAR_3188|M.marinum_M WDQE---KAFERLQSEHSRVLESFPTAVR-LSAGMSDDLEIAVKHGSTCV
MUL_3496|M.ulcerans_Agy99 WDQE---KAFERLQSEHNRVLESFPTAVR-LSAGMSDDLEIAVKHGSTCV
Mb2172c|M.bovis_AF2122/97 WDPD---EAFDRLQSEHNRVRAMFPHAIG-LSAGMSNDLEVAVKHGSTCV
Rv2148c|M.tuberculosis_H37Rv WDPD---EAFDRLQSEHNRVRAMFPHAIG-LSAGMSNDLEVAVKHGSTCV
MLBr_00919|M.leprae_Br4923 WNPD---QAFEQLRLEHRRVLRSHPDAIG-LSAGMSNDFEIAVKHGSTCV
MAV_2342|M.avium_104 SDPD---EAFDRLQSEHRRVLETHRNAVG-LSAGMSDDFEIAVKHGSTCV
Mflv_2993|M.gilvum_PYR-GCK ADPG---EAFARLRQERDRVQRDYEQRLG-LSAGMSGDLEAAVRHGSTCV
Mvan_3518|M.vanbaalenii_PYR-1 ADAG---EAFARLERERDRVQRRFQQRLG-LSAGMSGDLEEAVRHGSTCV
TH_2376|M.thermoresistible__bu WDAE---EAFARLAAEHARVQEGYSHRLG-LSAGMSGDLEAAVKHGSTCV
MAB_2011|M.abscessus_ATCC_1997 ADPD---QAFRRLQQEHERVRDRYPQANR-LSAGMSNDLEAAIKYGSTCV
MSMEG_2032|M.smegmatis_MC2_155 TDPERVRPCFRLLRSLRDQARDLDLIGPGELSMGMSGDYELAIAEGSTCV
: .* * : :. ** ***.* * *: *****
MMAR_3188|M.marinum_M RVGTALLGSRRLRSP------------------------------
MUL_3496|M.ulcerans_Agy99 RVGTALLGSRRLRSP------------------------------
Mb2172c|M.bovis_AF2122/97 RVGTALLGPRRLRSP------------------------------
Rv2148c|M.tuberculosis_H37Rv RVGTALLGPRRLRSP------------------------------
MLBr_00919|M.leprae_Br4923 RVGTALLGPSRLRSP------------------------------
MAV_2342|M.avium_104 RVGTALLGRRPLPSP------------------------------
Mflv_2993|M.gilvum_PYR-GCK RVGTALMGSRPLTSPEVVTRVTPSSQTTPPLDFPESASPQEGSEQ
Mvan_3518|M.vanbaalenii_PYR-1 RVGTALLGSRPLTSPEVVTPVTPSSQTPDSPELPESSPP-EGSSQ
TH_2376|M.thermoresistible__bu RVGTALLGPRPLTSPAVVTPVTPSSQTPPITRASEGSRD-EHTA-
MAB_2011|M.abscessus_ATCC_1997 RVGTALMGQRPLTSR------------------------------
MSMEG_2032|M.smegmatis_MC2_155 RVGQAIFGARPTPDSQYWPGN------------------------
*** *::* .