For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAHLSTPYLRDMGAKAADLVASSLTPAAAAYLNQPVDHLASLPWPFADDVTSFRYTVNVDPARVPRPTR AGEWGRHIVDLGGADYPVNMAERRHVLDTDPQRVKVRPGMELACWDLLIYYLRDLAQSYPDLLHLDEDGD HFHWRNDLLGTDQHFVLGDTGTLPGGPLFFLAAEIPDDLLLVIERDGRLYFDAGAVTFAAAWSASFDIGM DMYEIHGPVPRMTGEGMTARAEQFLKRLPANQVYRRLNWNLAASPTRTFDISLETLPDWGTHMPRALRDG DVSQVQFRIELEHFIRLPMTGAVTFNIRTFMASLEEIRAIPEYAEQLAAIIEELPDDIATYKGFADYQDD VVAYLRR
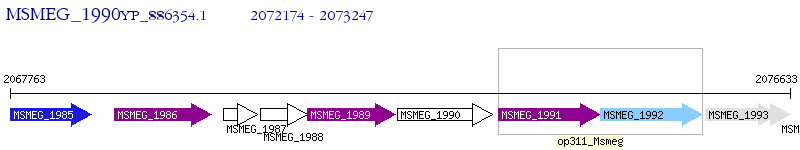
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1990 | - | - | 100% (357) | hypothetical protein MSMEG_1990 |
| M. smegmatis MC2 155 | MSMEG_4637 | - | 2e-74 | 42.77% (325) | hypothetical protein MSMEG_4637 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2624 | - | 7e-76 | 43.17% (322) | hypothetical protein Mflv_2624 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3957 | - | 1e-75 | 43.17% (322) | hypothetical protein Mvan_3957 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2624|M.gilvum_PYR-GCK ---MISAP-----------------------------DLIETFPFPFAAD
Mvan_3957|M.vanbaalenii_PYR-1 ---MISAP-----------------------------DLIETFPFPFAAD
MSMEG_1990|M.smegmatis_MC2_155 MTAHLSTPYLRDMGAKAADLVASSLTPAAAAYLNQPVDHLASLPWPFADD
:*:* * : ::*:*** *
Mflv_2624|M.gilvum_PYR-GCK T--YRYTTNVEPAGSSVHTPAGRWGERVVDID-SEYETELAERAEILAAD
Mvan_3957|M.vanbaalenii_PYR-1 E--YRYSTNVEPAGAAVPTPAGRWGERVVDLD-SEYETELAERAAILAAD
MSMEG_1990|M.smegmatis_MC2_155 VTSFRYTVNVDPARVPRPTRAGEWGRHIVDLGGADYPVNMAERRHVLDTD
:**:.**:** . * **.**.::**:. ::* .::*** :* :*
Mflv_2624|M.gilvum_PYR-GCK PTRYAVLPHMRVACWDAMLTLMREMSSAYPDTMSLERHGAGWRWRNDRLG
Mvan_3957|M.vanbaalenii_PYR-1 PTRYAVLPHMRPACWDAMLTLMRAMASAYPDTMSLQRHGTGWRWRNDRLG
MSMEG_1990|M.smegmatis_MC2_155 PQRVKVRPGMELACWDLLIYYLRDLAQSYPDLLHLDEDGDHFHWRNDLLG
* * * * *. **** :: :* ::.:*** : *:..* ::**** **
Mflv_2624|M.gilvum_PYR-GCK ITQDFVIGDETTLPAEPLAYIGGQVQEDIVLLDQRDGDLFGDAGVVTFAA
Mvan_3957|M.vanbaalenii_PYR-1 LAQDFVIGDEASLPAEPLAYIAGQVQEDIVLLDQRDGDLFGDAGVVTFAA
MSMEG_1990|M.smegmatis_MC2_155 TDQHFVLGDTGTLPGGPLFFLAAEIPDDLLLVIERDGRLYFDAGAVTFAA
*.**:** :**. ** ::..:: :*::*: :*** *: ***.*****
Mflv_2624|M.gilvum_PYR-GCK DWSFGFDVGMTFLEIHGPVPRLRQTGVITRAREFLMRLQPGETYRRTNWT
Mvan_3957|M.vanbaalenii_PYR-1 DWSFGFDVGMTFLEIHGPVPRLRETGVITRAREFLMRLQPGETYRRTNWS
MSMEG_1990|M.smegmatis_MC2_155 AWSASFDIGMDMYEIHGPVPRMTGEGMTARAEQFLKRLPANQVYRRLNWN
** .**:** : ********: *: :**.:** ** ..:.*** **.
Mflv_2624|M.gilvum_PYR-GCK LTID--RRLDVSTERYCEWGPDRTLIQTVPDDEFGRRVHLRVEVQHLIRL
Mvan_3957|M.vanbaalenii_PYR-1 LTIG--RRLDVSTELYHEWGPDRTAIQTVSDEEFGRLVHLRVEVQHLIRL
MSMEG_1990|M.smegmatis_MC2_155 LAASPTRTFDISLETLPDWG--THMPRALRDGDVS-QVQFRIELEHFIRL
*: . * :*:* * :** ::: * :.. *::*:*::*:***
Mflv_2624|M.gilvum_PYR-GCK PESGSICFLIRSYMLPLADLAAFEPWRQRAAAVLAELPDDIADYKGIIAY
Mvan_3957|M.vanbaalenii_PYR-1 PESGSICFLIRTYMLPFADLAAFEPWRQRAAAVLAELPDDMADYKGIIAY
MSMEG_1990|M.smegmatis_MC2_155 PMTGAVTFNIRTFMASLEEIRAIPEYAEQLAAIIEELPDDIATYKGFADY
* :*:: * **::* .: :: *: : :: **:: *****:* ***: *
Mflv_2624|M.gilvum_PYR-GCK RDRAVAWLRNAG
Mvan_3957|M.vanbaalenii_PYR-1 RDRAVAWLRNAG
MSMEG_1990|M.smegmatis_MC2_155 QDDVVAYLRR--
:* .**:**.