For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGMRPTARMPKLTRRSRVLIAFALVAVLLLLLGPRLIDTYVDWLWFGELGYRSVFTTVLATRLIVFVVVA LAIGAIVFAGLALAYRTRPVFVPTAGPNDPVARYRTTVMARLRLFGIGVPVFIGLLAGIVAQSYWVKIQL FLHGGDFGITDPEFGKDLGFYAFDLPFYRLVLTYLFVATFLAFVANLLGHYLFGGIRLTGRVGALSRAAR IQLISLAGTLIVLKAFAYWLDRYELLSNDRSAKPFTGAGYTDINAVLPAKLIMLAIAVICAVAVFSALVL RDLRIPAIGVALLLLSSLVVGAGWPLIVEQFSVKPNAAQKEAEYISRSIEATRHAYGLTDETVTYRNYEN TGQTTAAQVAADRATTSNIRLLDPTIVSPAFTQFQQGKNFYYFPDQLSIDRYIGPDGNLRDYVVAARELN PDRLIDNQRDWINRHTVYTHGNGFIASPANTVRGVANDPNQNGGYPEFLASVVGANGSVISPGPAPLDQP RIYFGPVISNTPADYAIVGKTGDTDREYDYETNTETKNYTYGGKGGVPIGNWLNRSVFAAKFAERNFLFS NVIGENSKILFNRDPAERVEAVAPWLTTDTSVYPAIVNKRMVWIVDGYTTLDNYPYSELTTLSSATADSN EVAVNRLAPDKKVSYIRNSVKATVDAYDGTVTLYAQDENDPVLKAWMDVFPGTVKPKADITPELQAHLRY PEDLFKVQRALLAKYHVDNPVTFFSAQDFWDVPLDPNPTASSFQPPYYIVAKDLVKNDNSASFQLTSALN RFQRDFLAAYVSASSDPETYGKLTVLTIPGQVNGPKLAFNAISTDTAVSQDLGVIGRDNQNRIRWGNLLT LPVADGGLLYVAPVYASPGSSDAASSYPRLIRVAMLYNDRVGYGPTVSDALTELFGPGAGATATDVAPAE GRPAQSTPNGQQPAASPPPAANADGRPAQAPPPPSAATPTGPVQISQAKAEALQDLESALTAAQEAQRSG DFAEYGQALQRLNDAMKKYDSAK
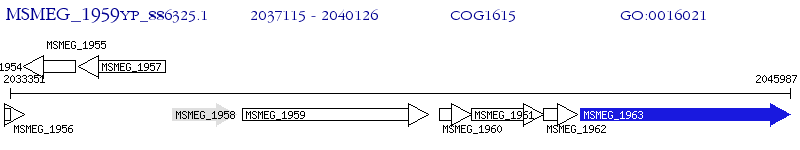
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1959 | - | - | 100% (1003) | hypothetical protein MSMEG_1959 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3215c | - | 0.0 | 79.34% (1002) | hypothetical protein Mb3215c |
| M. gilvum PYR-GCK | Mflv_4654 | - | 0.0 | 80.06% (1003) | hypothetical protein Mflv_4654 |
| M. tuberculosis H37Rv | Rv3193c | - | 0.0 | 79.34% (1002) | hypothetical protein Rv3193c |
| M. leprae Br4923 | MLBr_00644 | - | 0.0 | 76.17% (1003) | hypothetical protein MLBr_00644 |
| M. abscessus ATCC 19977 | MAB_3498c | - | 0.0 | 69.70% (1010) | hypothetical protein MAB_3498c |
| M. marinum M | MMAR_1371 | - | 0.0 | 79.82% (1006) | transmembrane protein |
| M. avium 104 | MAV_4137 | - | 0.0 | 79.46% (1003) | hypothetical protein MAV_4137 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2505 | - | 0.0 | 79.42% (1006) | hypothetical protein MUL_2505 |
| M. vanbaalenii PYR-1 | Mvan_1814 | - | 0.0 | 81.63% (1007) | hypothetical protein Mvan_1814 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4654|M.gilvum_PYR-GCK MGMRPAARMPSLTQRSRFLIAVSAVLVLLLLLGPRFIDTYVDWLWFGELG
Mvan_1814|M.vanbaalenii_PYR-1 --MRPAARMPNLTRRSRVMIAVALAVVVLLLLGPRLVDTYVNWLWFGELG
MSMEG_1959|M.smegmatis_MC2_155 MGMRPTARMPKLTRRSRVLIAFALVAVLLLLLGPRLIDTYVDWLWFGELG
MMAR_1371|M.marinum_M MGMRPTARMPKLTRRSRVLILIALGVIALLLAGPRLIDAYVDWLWFGELG
MUL_2505|M.ulcerans_Agy99 MGMRPTARMPKLTRRSRVLILIALGVIALLLAGPRLIDAYVDWLWFGELG
MAV_4137|M.avium_104 MGMRPTARMPKLTRRSRILILIALGVIALLLAGPRLIDAYVDWLWFGELG
Mb3215c|M.bovis_AF2122/97 MGMRSAARMPKLTRRSRILIMIALGVIVLLLAGPRLIDAYVDWLWFGELG
Rv3193c|M.tuberculosis_H37Rv MGMRSAARMPKLTRRSRILIMIALGVIVLLLAGPRLIDAYVDWLWFGELG
MLBr_00644|M.leprae_Br4923 MGMRPAARMPKLTRRSRTLIMVALGVIVLLLAGPRLVDAYVEWLWFGELG
MAB_3498c|M.abscessus_ATCC_199 MGMRPNGALPRLTRRSRRLVAASLVIVVLLLIGPRLVDTYINWLWFGELG
**. . :* **:*** :: : : *** ***::*:*::********
Mflv_4654|M.gilvum_PYR-GCK YRSVFTTQIITRLVIFFVVAILVGAVVFAGLALAYRTRPVFVPTAGPNDP
Mvan_1814|M.vanbaalenii_PYR-1 YRSVFTTQIVTRLLLFLAVAVVFGAVVFAAMALAYRTRPVFVPTAGPNDP
MSMEG_1959|M.smegmatis_MC2_155 YRSVFTTVLATRLIVFVVVALAIGAIVFAGLALAYRTRPVFVPTAGPNDP
MMAR_1371|M.marinum_M YRSVFTTVLVTRFLVFLVAGVLVGGIVFAGLALAYRTRPVFVPNND-NDP
MUL_2505|M.ulcerans_Agy99 YLSVFTTVLVTRFLVFLVAGVLVGGIVFAGLALAYRTRPVFVPNND-NDP
MAV_4137|M.avium_104 YRSVFSTVLVTRFVVFLIAGLLVGGIVFAGLAVAYRTRPVFVPSND-NDP
Mb3215c|M.bovis_AF2122/97 YRSVFTTMLATRIVVCLVAGVVVGGIVFGGLALAYRTRPVFVPDAD-NDP
Rv3193c|M.tuberculosis_H37Rv YRSVFTTMLATRIVVCLVAGVVVGGIVFGGLALAYRTRPVFVPDAD-NDP
MLBr_00644|M.leprae_Br4923 YRSVFSTVLVTRVVVFLVAGLVVGGIVFAGLAVAYRTRPVFVPSHD-NDP
MAB_3498c|M.abscessus_ATCC_199 FRGVFTTVLLTRLALFLIVGTLVAAVVFAGFGLAYRARPVFVPAKGPGDA
: .**:* : **. : . .. ...:**..:.:***:****** . .*.
Mflv_4654|M.gilvum_PYR-GCK IARYRTAVMARLRLIGIGVPLAIGVLAGFVGQSYWQRVQLFLHGGDFGVS
Mvan_1814|M.vanbaalenii_PYR-1 IARYRTAVMARLRLVGIGVPVAVGLLAGLIAQNYWQRVQLFLHGGSFGVS
MSMEG_1959|M.smegmatis_MC2_155 VARYRTTVMARLRLFGIGVPVFIGLLAGIVAQSYWVKIQLFLHGGDFGIT
MMAR_1371|M.marinum_M VARYRTVVLARLRLFGIGIPAAIGLLAGIVAQSYWVRIQLFLHGGDFGIT
MUL_2505|M.ulcerans_Agy99 VARYRTVVLARLRLFGIGIPAAIGLLAGIVAQSYWVRIQLFLHGGDFGIT
MAV_4137|M.avium_104 VARYRALVLSRLRLVSIGVPVAIGLLAGIIAQSYWVRIQLFLHGGDFGIK
Mb3215c|M.bovis_AF2122/97 VARYRAVVLARLRLVGIGIPAAIGLLAGIVAQSYWARIQLFLHGGDFGVR
Rv3193c|M.tuberculosis_H37Rv VARYRAVVLARLRLVGIGIPAAIGLLAGIVAQSYWARIQLFLHGGDFGVR
MLBr_00644|M.leprae_Br4923 VARYRAMALSRLRLIGVGIPAAIGLLAGIIAQSYWVRIQLFLHGDNFGIR
MAB_3498c|M.abscessus_ATCC_199 LAQYRALILSRVRLFLIGVPILIGVLAGVVAQSYWMPVQLFLEGGDFGIK
:*:**: ::*:**. :*:* :*:***.:.*.** :****.*..**:
Mflv_4654|M.gilvum_PYR-GCK DPQFGIDLGFYAFDLPFYRLVLTYLFVGVFLAFLANLLGHYLFGGIRLAG
Mvan_1814|M.vanbaalenii_PYR-1 DPQFGIDLGFYAFDLPFYRLMLTYLFAATFLAFIANLLGHYLFGGIRLAG
MSMEG_1959|M.smegmatis_MC2_155 DPEFGKDLGFYAFDLPFYRLVLTYLFVATFLAFVANLLGHYLFGGIRLTG
MMAR_1371|M.marinum_M DPQFGKDLGFYAFELPFYRLLLSYLFVAIFLAFVANVVSHYLFGGIRLTG
MUL_2505|M.ulcerans_Agy99 DPQFGKDLGFYAFELPFYRLLLSYLFVAIFLAFVANVVSHYLFGGIRLTG
MAV_4137|M.avium_104 DPQFGKDLGFYAFELPFYRLLLSYLFVAVFLAFVANLLAHYIFGGIRLSG
Mb3215c|M.bovis_AF2122/97 DPQFGRDLGFYAFELPFYRLMLSYMLVSVFLAFVANLVAHYIFGGIRLSG
Rv3193c|M.tuberculosis_H37Rv DPQFGRDLGFYAFELPFYRLMLSYMLVSVFLAFVANLVAHYIFGGIRLSG
MLBr_00644|M.leprae_Br4923 DPQFGKDLGFYAFELPFYRLVLSYVFVAVFLAFVVNLLAHYIFGGIRLSG
MAB_3498c|M.abscessus_ATCC_199 DPQFGLDLGFFAFDLPFYRFVLTYLFIAASIALIVNGLVHYIFGGIRLSG
**:** ****:**:*****::*:*:: . :*::.* : **:******:*
Mflv_4654|M.gilvum_PYR-GCK RSGALSRAARIQLITLVGLLMLLKAVAYWFDRYELLSHTRGGKPFTGAGY
Mvan_1814|M.vanbaalenii_PYR-1 RSGALSRAARIQLIALVGFLMLLKAVAYWLDRYELLSHTRGGKPFTGAGY
MSMEG_1959|M.smegmatis_MC2_155 RVGALSRAARIQLISLAGTLIVLKAFAYWLDRYELLSNDRSAKPFTGAGY
MMAR_1371|M.marinum_M RSGALSRSARIQLVSLVGVLVLLKAVAYWLDRYELLSHTRGGKPFTGAGY
MUL_2505|M.ulcerans_Agy99 RSGALSRSARIQLVSLVGVLVLLKTVAYWLNRYELLSHTRGGKPFTGAGY
MAV_4137|M.avium_104 RTGALSRSARIQLVTLVGLLVLLKAVAYWLDRYELLSHTRGGKPFTGAGY
Mb3215c|M.bovis_AF2122/97 RTGALSRSARVQLVSLVGVLVLLKAVAYWLDRYELLSHTRGGKPFTGAGY
Rv3193c|M.tuberculosis_H37Rv RTGALSRSARVQLVSLVGVLVLLKAVAYWLDRYELLSHTRGGKPFTGAGY
MLBr_00644|M.leprae_Br4923 RTGALSRLARLQLVSLVGVLVLLKAVAYWLDRYELLSRTRSSKPFTGAGY
MAB_3498c|M.abscessus_ATCC_199 RSGTLSRPARIQLITFAGILVLLKVAAYWLDRYELLSHTRAGKPFTGAGY
* *:*** **:**:::.* *::**. ***::******. *..********
Mflv_4654|M.gilvum_PYR-GCK TDINAVLPAKLILMAIAVICAVAVFSAIFLRDLRIPAIGVVLLLLSSLVV
Mvan_1814|M.vanbaalenii_PYR-1 TDINAVLPAKLILMVIAVICAAAVFSAIVLRDLRIPAIGVVLLLLSSLIV
MSMEG_1959|M.smegmatis_MC2_155 TDINAVLPAKLIMLAIAVICAVAVFSALVLRDLRIPAIGVALLLLSSLVV
MMAR_1371|M.marinum_M TDINAVLPAKLILMAIAVICAAAVFSAIVLRDLRIPAIGLVLLLLSSLIV
MUL_2505|M.ulcerans_Agy99 TDINAVLPAKLILMAIAVICAAAVFSAIVLRDLRIPAIGLVLLLLSSLIV
MAV_4137|M.avium_104 TDINAVLPAKLILMAIALICAAAVFSAITMRDLRIPAIGLVLLLLSSLIV
Mb3215c|M.bovis_AF2122/97 TDINAVLPAKLILMAIALICAAAVFSAIALRDLRIPAIGLVLLLLSSLIV
Rv3193c|M.tuberculosis_H37Rv TDINAVLPAKLILMAIALICAAAVFSAIALRDLRIPAIGLVLLLLSSLIV
MLBr_00644|M.leprae_Br4923 TDINAVQPAKLILIAIALICAAAVFSAITLRDFRIPAIGLVLLMLSSLIV
MAB_3498c|M.abscessus_ATCC_199 TDINAVLPAKLILLAIAVICAVAVFSALVLKDLRIPAIGLALLLLSSLVV
****** *****::.**:***.*****: ::*:******:.**:****:*
Mflv_4654|M.gilvum_PYR-GCK GAGWPLVVEQISVRPNAAQKESEYISRSITATRQAYGLTEESVTYRDYPG
Mvan_1814|M.vanbaalenii_PYR-1 GAGWPLVVEQISVRPNAAQKESEYISRSITATRQAYGLTDEAVEYRDYPG
MSMEG_1959|M.smegmatis_MC2_155 GAGWPLIVEQFSVKPNAAQKEAEYISRSIEATRHAYGLTDETVTYRNYEN
MMAR_1371|M.marinum_M GAAWPMIVEQISVKPNAAQKESEYISRSITATRQAYGLTSNVVTYRNYTG
MUL_2505|M.ulcerans_Agy99 GAAWPMIVEQISVKPNAAQKESEYISRSITATRQAYGLTSNVVTYRNYTG
MAV_4137|M.avium_104 GAGWPLIVEQISVKPNAAQKESEYISRSITATRQAYGLTSDVVTYRNYTG
Mb3215c|M.bovis_AF2122/97 GAGWPLIVEQISVKPNAAQKESEYISRSITATRQAYGLTSDVVTYRNYSG
Rv3193c|M.tuberculosis_H37Rv GAGWPLIVEQISVKPNAAQKESEYISRSITATRQAYGLTSDVVTYRNYSG
MLBr_00644|M.leprae_Br4923 GTGWPLIVEQLIAKPDAVRKESEYIRRSITATRHAYGLTEDVVTYRNYIG
MAB_3498c|M.abscessus_ATCC_199 GAGWPLIVEQFSVKPNAAQKESEYIARSIKATRDAYGLTDDVVTYRDYSG
*:.**::***: .:*:*.:**:*** *** ***.*****.: * **:* .
Mflv_4654|M.gilvum_PYR-GCK NA------SATAQQVAADRATTSNIRVLDPNIVAPAFTQFQQGKNFYFFP
Mvan_1814|M.vanbaalenii_PYR-1 NA------TATAQQVAADRATTSNIRVLDPNIVSPAFTQFQQGKNFYFFP
MSMEG_1959|M.smegmatis_MC2_155 TG------QTTAAQVAADRATTSNIRLLDPTIVSPAFTQFQQGKNFYYFP
MMAR_1371|M.marinum_M DG------EATAQQVAADRATTSNIRLLDPTIVSPAFTQFQQGKNFYYFP
MUL_2505|M.ulcerans_Agy99 DG------EATAQQVAADRATTSNIRLLDPTIVSPAFTQFQQGKNFYYFP
MAV_4137|M.avium_104 DA------QATAQQVADDRATTSNIRLLDPTIVSPAFTQFQQGKNFYYFP
Mb3215c|M.bovis_AF2122/97 DS------PATAQQVAADRATTSNIRLLDPTIVSPAFTQFQQGKNFYYFP
Rv3193c|M.tuberculosis_H37Rv DS------PATAQQVAADRATTSNIRLLDPTIVSPAFTQFQQGKNFYYFP
MLBr_00644|M.leprae_Br4923 DA------PAIAQQIATDHATTSNIRLLDPTIVSPAFTQFQRGKNFYYFP
MAB_3498c|M.abscessus_ATCC_199 TASSPAGSQQLAKQVAADRSTIANIRVLDPNIISPAFTQLQQGKNFYAFP
. * *:* *::* :***:***.*::*****:*:***** **
Mflv_4654|M.gilvum_PYR-GCK DQLNMDRYRDDSGNLRDYVVAARELNPDRLIDNQRDWINRHTVFTHGNGF
Mvan_1814|M.vanbaalenii_PYR-1 DQLNMDRYRDEDGNLRDYVVAARELNPDRLIDNQRDWINRHSVYTHGNGF
MSMEG_1959|M.smegmatis_MC2_155 DQLSIDRYIGPDGNLRDYVVAARELNPDRLIDNQRDWINRHTVYTHGNGF
MMAR_1371|M.marinum_M DQLSIDRYFDRNNNLRDYVVAARELNPDRLIDNQRDWINRHTVYTHGNGF
MUL_2505|M.ulcerans_Agy99 DQLSIDRYFDRNNNLRDYVVAARELNPDRLIDNQRDWINRHTVYTHGNGF
MAV_4137|M.avium_104 DQLSIDRYLDRNGALRDYVVAARELNPDRLIDNQRDWINRHTVYTHGNGF
Mb3215c|M.bovis_AF2122/97 DQLSIDRYLDRNGNLRDYVVAARELNPDRLIDNQRDWINRHTVYTHGNGF
Rv3193c|M.tuberculosis_H37Rv DQLSIDRYLDRNGNLRDYVVAARELNPDRLIDNQRDWINRHTVYTHGNGF
MLBr_00644|M.leprae_Br4923 DQLSIDRYLDKKGNLRDYVVAARELNPDRLIDNQRDWINRHTVYTHGNGF
MAB_3498c|M.abscessus_ATCC_199 DALSIDRYQDKNGSLRDYVVAARELDPAKLRDNQRDWINRHTVYTHGNGF
* *.:*** . .. ***********:* :* **********:*:******
Mflv_4654|M.gilvum_PYR-GCK IASPANTVRGIANDPNQNGGYPEFLASVVGANGEVSSPGPAPLAQPRIYF
Mvan_1814|M.vanbaalenii_PYR-1 IASPANTVRGIANDPNQNGGYPEFLASVVGANGEVVSPGPAPLDQPRIYF
MSMEG_1959|M.smegmatis_MC2_155 IASPANTVRGVANDPNQNGGYPEFLASVVGANGSVISPGPAPLDQPRIYF
MMAR_1371|M.marinum_M IASPANTVRGIANDPNQNGGYPEFLVNVVGANGTVVSDGPAPLDQPRIYY
MUL_2505|M.ulcerans_Agy99 IASPANTVRGIANDPNQNGGYPEFLVNVVGANGTVVSDGPAPLDQPRIYY
MAV_4137|M.avium_104 IASPANTVRGIANDPNQNGGYPEFLVNVVGANGNVVSDGPAPLDQPRVYF
Mb3215c|M.bovis_AF2122/97 IASPANTVRGIANDPNQNGGYPEFLVNVVGANGTVVSDGPAPLDQPRIYF
Rv3193c|M.tuberculosis_H37Rv IASPANTVRGIANDPNQNGGYPEFLVNVVGANGTVVSDGPAPLDQPRIYF
MLBr_00644|M.leprae_Br4923 IASPANTVRGIANDPNQNGGYPQFLVNVVG-NGTVVSEGPARLDQPRVYF
MAB_3498c|M.abscessus_ATCC_199 IAAPANTVRGVADKPDENGGYPEFLVNAVDDNGKVLSDGPAPLAQPRVYY
**:*******:*:.*::*****:**...*. ** * * *** * ***:*:
Mflv_4654|M.gilvum_PYR-GCK GPVIASAADDYAIVGESG-TPREYDYETNTDTRNYTYTGSGGVPIGNWLT
Mvan_1814|M.vanbaalenii_PYR-1 GPVIANTPADYAIVGESG-TPREYDYETNTATRNYTYTGSGGVPIGNWLT
MSMEG_1959|M.smegmatis_MC2_155 GPVISNTPADYAIVGKTGDTDREYDYETNTETKNYTYGGKGGVPIGNWLN
MMAR_1371|M.marinum_M GPVISNTPADYAIVGKTG-ADREYDYETSADTKNYTYTGSGGVSVGSWIS
MUL_2505|M.ulcerans_Agy99 GPVISNTPADYAIVGKTG-ADREYDYETSADTKNYTYTGSGGVSVGSWIS
MAV_4137|M.avium_104 GPVISNTSADYAIVGRNG-ADREYDYETSTETKNYTYTGLGGVPIGDWLS
Mb3215c|M.bovis_AF2122/97 GPVISNTSADYAIVGRNG-DDREYDYETNIDTKRYTYTGSGGVPLGGWLA
Rv3193c|M.tuberculosis_H37Rv GPVISNTSADYAIVGRNG-DDREYDYETNIDTKRYTYTGSGGVPLGGWLA
MLBr_00644|M.leprae_Br4923 GPVISNTSADYAIVGKNG-DDREYDYETNTDTKRYTYAGSGGVPIGSWLS
MAB_3498c|M.abscessus_ATCC_199 GPIIASDTNDYAIVGKNG-NDREYDYENNAGTKNSTYTGSGGVPVGGALA
**:*:. . ******..* ******.. *:. ** * ***.:*. :
Mflv_4654|M.gilvum_PYR-GCK RSVFAAKYAERNFLFSNVINENSKILFNRDPADRVEAVAPWLTTDTTVYP
Mvan_1814|M.vanbaalenii_PYR-1 RSVFAAKYAERNFLFSNVIGENSKILFNRDPADRVEAVAPWLTTDTAVYP
MSMEG_1959|M.smegmatis_MC2_155 RSVFAAKFAERNFLFSNVIGENSKILFNRDPAERVEAVAPWLTTDTSVYP
MMAR_1371|M.marinum_M RTVFAAKFAERNFLFSNVIGSNSKILFNRDPAQRVEAVAPWLTTDSAVYP
MUL_2505|M.ulcerans_Agy99 RTVFAAKFAERNFLFSNVIGSNSKILFNRDPAQRVEAVAPWLTTDSAVYP
MAV_4137|M.avium_104 RSVFAAKFAERNFLFSNVIGSNSKILFNRDPARRVEAVAPWLTTDSSVYP
Mb3215c|M.bovis_AF2122/97 RSVFAAKFAERNFLFSNVIGSNSKILFNRDPAQRVEAVAPWLTTDSAVYP
Rv3193c|M.tuberculosis_H37Rv RSVFAAKFAERNFLFSNVIGSNSKILFNRDPAQRVEAVAPWLTTDSAVYP
MLBr_00644|M.leprae_Br4923 RSVFAAKFAERNFLFSSVIGSNSKILFNRDPAQRVEAVAPWLTTDSSVYP
MAB_3498c|M.abscessus_ATCC_199 RTVFGLKYAERNFLFSNVIGDNSKILFNRDPSRRVEAVAPWLTVDSGTYP
*:**. *:********.**..**********: **********.*: .**
Mflv_4654|M.gilvum_PYR-GCK AIVNERIVWIVDGYTTLDNYPYSELSTLSSVTTDSNEVAQNRLQLDKQVS
Mvan_1814|M.vanbaalenii_PYR-1 AIVNKRIVWIVDGYTTLDNYPYSELMSLSSATTDSNEVALNRLQPDKQVS
MSMEG_1959|M.smegmatis_MC2_155 AIVNKRMVWIVDGYTTLDNYPYSELTTLSSATADSNEVAVNRLAPDKKVS
MMAR_1371|M.marinum_M AIVNKRMVWILDGYTTLDNYPYSQLTSLSSATADSNEVAFNRLLPDKQVS
MUL_2505|M.ulcerans_Agy99 AIVNKRMVWILDGYTTLDNYPYSQLTSLSSATADSNEVAFNRLLPDKQVS
MAV_4137|M.avium_104 AIVNKRLVWIIDGYTTLDNYPYSELTSLESATADSNEVAFNKLAPDKRVS
Mb3215c|M.bovis_AF2122/97 AIVNKRLVWIVDGYTTLDNYPYSELTSLSSATADSNEVAFNRLVPDKKVS
Rv3193c|M.tuberculosis_H37Rv AIVNKRLVWIVDGYTTLDNYPYSELTSLSSATADSNEVAFNRLVPDKKVS
MLBr_00644|M.leprae_Br4923 AIVNKRLVWIIDGYTTLDNYPYSERTSLSSATADSTEVAFNRLAPDKRVA
MAB_3498c|M.abscessus_ATCC_199 AIVDKRLVWIVDGYTTLDNYPYSQQTSLSEATFDSQVGRTGGALPNQQVS
***::*:***:************: :*...* ** . :::*:
Mflv_4654|M.gilvum_PYR-GCK YIRNSVKATVDAYDGTVTLYAQDEQDPVLQAWMKVFPDSVKPKSDITPEL
Mvan_1814|M.vanbaalenii_PYR-1 YIRNSVKATVDAYDGTVTLYAQDEQDPVLQAWMKVFPDTVKPKADITPEL
MSMEG_1959|M.smegmatis_MC2_155 YIRNSVKATVDAYDGTVTLYAQDENDPVLKAWMDVFPGTVKPKADITPEL
MMAR_1371|M.marinum_M YIRNSVKATVDAYDGTVTLYQQDEQDPVLKAWMQVFPGTVKPKGDISPEL
MUL_2505|M.ulcerans_Agy99 YIRNSVKATVDAYDGTVTLYQQDEQDPVLKAWMQVFPGTVKPKGDISPEL
MAV_4137|M.avium_104 YIRNSVKATVDAYDGTVTLYQQDEQDPVLKAWMQVFPGTVKPKSDISPEL
Mb3215c|M.bovis_AF2122/97 YIRNSVKATVDAYDGTVTLYQQDEKDPVLKAWMQVFPGTVKPKSDIAPEL
Rv3193c|M.tuberculosis_H37Rv YIRNSVKATVDAYDGTVTLYQQDEKDPVLKAWMQVFPGTVKPKSDIAPEL
MLBr_00644|M.leprae_Br4923 YIRNSVKATVDAYDGAVTLYQQDEYDPVLKVWMKVFPGTVRPKGDITPEL
MAB_3498c|M.abscessus_ATCC_199 YIRNSVKATVDAYDGTVTLYQQDEKDPVLKAWMKIFPGTVKPKADISDDL
***************:**** *** ****:.**.:**.:*:**.**: :*
Mflv_4654|M.gilvum_PYR-GCK QEHLRYPEDLFKVQRALLAKYHVDDPVTFFSTSDFWDVPLDPNPTASSYQ
Mvan_1814|M.vanbaalenii_PYR-1 QEHLRYPEDLFKVQRALLAKYHVDDPVTFFSTSDFWDVPLDPNPTASSYQ
MSMEG_1959|M.smegmatis_MC2_155 QAHLRYPEDLFKVQRALLAKYHVDNPVTFFSAQDFWDVPLDPNPTASSFQ
MMAR_1371|M.marinum_M AAHLRYPEDLFKVQRMLLAKYHVNDPVTFFSTSDFWDVPLDPNPTASSYQ
MUL_2505|M.ulcerans_Agy99 AAHLRYPEDLFKVQRMLLAKYHVNDPVTFFSTSDFWDVPLDPNPTASSYQ
MAV_4137|M.avium_104 AEHLRYPEDLFKVQRMLLAKYHVNDPVTFFSTSDFWDVPLDPNPTASSYQ
Mb3215c|M.bovis_AF2122/97 AEHLRYPEDLFKVQRMLLAKYHVNDPVTFFSTSDFWDVPLDPNPTASSYQ
Rv3193c|M.tuberculosis_H37Rv AEHLRYPEDLFKVQRMLLAKYHVNDPVTFFSTSDFWDVPLDPNPTASSYQ
MLBr_00644|M.leprae_Br4923 AEHLRYPEDLFKVQRMLLAKYHVNDPGTFFNTSDFWDVPLDPNPTASSYQ
MAB_3498c|M.abscessus_ATCC_199 KRHLRYPEDLFKVQRTLLARYHVNDPVTFFSTSDFWQVPDDPNAPTGS-Q
************* ***:***::* ***.:.***:** ***..:.* *
Mflv_4654|M.gilvum_PYR-GCK PPYYIVAKDLAENNGSASFQLTSAMNRFRRDFLAAYISASSDPETYGRLT
Mvan_1814|M.vanbaalenii_PYR-1 PPYYIVAKDLAENNNSSSFQLTSAMNRFRRDFLAAYISASSDPETYGKLT
MSMEG_1959|M.smegmatis_MC2_155 PPYYIVAKDLVKNDNSASFQLTSALNRFQRDFLAAYVSASSDPETYGKLT
MMAR_1371|M.marinum_M PPYYIVAKNIAKNDNSASYQLISAMNRFKRDYLAAYISASSDPATYGKIT
MUL_2505|M.ulcerans_Agy99 PPYYIVAKNIAKNDNSASYQLISAMNRFKRDYLAAYISASSDPATYGKIT
MAV_4137|M.avium_104 PPYYIVAKNIAKNDNSSSYQLTSAMNRFKRDYLAAYISASSDPATYGRIT
Mb3215c|M.bovis_AF2122/97 PPYYIVAKNIAKDDNSASYQLISAMNRFKRDYLAAYISASSDPATYGNLT
Rv3193c|M.tuberculosis_H37Rv PPYYIVAKNIAKDDNSASYQLISAMNRFKRDYLAAYISASSDPATYGNLT
MLBr_00644|M.leprae_Br4923 PPYYIVAKNILKNDRSASYQLTSAMNRFKQDFLAAYISASSDPETYGKIT
MAB_3498c|M.abscessus_ATCC_199 PPYYIVAKDITKNDNSASFQLTSALNRFQRDFLAAYVSASSDPETYGKIT
********:: ::: *:*:** **:***::*:****:****** ***.:*
Mflv_4654|M.gilvum_PYR-GCK VLTVPGQVNGPKLAFNAISTDTAISTELGQIGRDGQNRIRWGNLLTLPMG
Mvan_1814|M.vanbaalenii_PYR-1 VLTIPGQVNGPKLAFNAISTDTAVSQDLGVIGRDNQNRIRWGNLLTLPMG
MSMEG_1959|M.smegmatis_MC2_155 VLTIPGQVNGPKLAFNAISTDTAVSQDLGVIGRDNQNRIRWGNLLTLPVA
MMAR_1371|M.marinum_M VLTIPGQVNGPKLANNAITTDPAVSQDLGVIGRDNQNRIRWGNLLTLPVG
MUL_2505|M.ulcerans_Agy99 VLTIPGQVNGPKLANNAITTDPAVSQDLGVIGRDNQNRIRWGNLLTLPVG
MAV_4137|M.avium_104 VLTIPGQVNGPKLANNAITTDPAVSQDLGVIGRDNQNRIRWGNLLTLPVG
Mb3215c|M.bovis_AF2122/97 VLTIPGQVNGPKLANNAITTDPAVSQDLGVIGRDNQNRIRWGNLLTLPVA
Rv3193c|M.tuberculosis_H37Rv VLTIPGQVNGPKLANNAITTDPAVSQDLGVIGRDNQNRIRWGNLLTLPVA
MLBr_00644|M.leprae_Br4923 VLTIPGNVNGPKLANNAITTDPAVSQDLGVIGRDNQNRIRWGNLLTLPVA
MAB_3498c|M.abscessus_ATCC_199 VLTVPGTVQGPKLVNNAITTDNQVSSHVGIIK--NQNILKWGNLLTLPVA
***:** *:****. ***:** :* .:* * .** ::********:.
Mflv_4654|M.gilvum_PYR-GCK DGGLLYVAPIYASPGNTDAASTYPRLIRVAMMYNDQIGYGPTVRDALTDL
Mvan_1814|M.vanbaalenii_PYR-1 QGGLLYVAPVYASPGASDAASSYPRLIRVAMMYNDQIGYGPTVRDALTDL
MSMEG_1959|M.smegmatis_MC2_155 DGGLLYVAPVYASPGSSDAASSYPRLIRVAMLYNDRVGYGPTVSDALTEL
MMAR_1371|M.marinum_M QGGLLYVEPVYASPGASDAASSYPRLIRVAMMYNDKIGYGPTVRDALNGL
MUL_2505|M.ulcerans_Agy99 QGGLLYVEPVYASPGASDAASSYPRLIRVAMMYNDKIGYGPTVRDALNGL
MAV_4137|M.avium_104 QGGLLYVEPVYASPGASDAASSYPRLIRVAMMYNDKIGYGPTVRDALTGL
Mb3215c|M.bovis_AF2122/97 QGGLLYVEPVYASPGASDAASSYPRLIRVAMMYNDKVGYGPTVRDALTGL
Rv3193c|M.tuberculosis_H37Rv RGGLLYVEPVYASPGASDAASSYPRLIRVAMMYNDKVGYGPTVRDALTGL
MLBr_00644|M.leprae_Br4923 QGGLLYVEPVYASPGVSDAASSYPRLIRVAMMYNDKIGYGPTVGDALTGL
MAB_3498c|M.abscessus_ATCC_199 NGGLLFVEPLYASPGQGDQSS-YPRLIRVGMYYNGKVGYATTVRDALDMV
****:* *:***** * :* *******.* **.::**..** *** :
Mflv_4654|M.gilvum_PYR-GCK FGPGADATATGPAATEPPAGQAPQTQGNNTAPPAAQPPNRQGQAPAGR-P
Mvan_1814|M.vanbaalenii_PYR-1 FGPGADATATGPAATEPPAGQAPQPQGNN-QPPAAAPPNRPGQAPTPQQP
MSMEG_1959|M.smegmatis_MC2_155 FGPGAGATATDVAPAE---GRPAQSTPNGQQPAASPPPAANADGRPAQAP
MMAR_1371|M.marinum_M FGPGAGDAATGIQPTEG--GAPANVPPNNAPSPEALPGTPP-------SP
MUL_2505|M.ulcerans_Agy99 FGPGAGDAATGIQPTEG--GAPANVPPNNAPSPEALPGTPP-------SP
MAV_4137|M.avium_104 FGPGAGAAATNIQPTEG--GAPAASPPANAPAPAVTPGS---------AP
Mb3215c|M.bovis_AF2122/97 FGPGAGATATGIAPTEA--AVPP-SPAANPPPPASGPQP---------PP
Rv3193c|M.tuberculosis_H37Rv FGPGAGATATGIAPTEA--AVPP-SPAANPPPPASGPQP---------PP
MLBr_00644|M.leprae_Br4923 FGPGAASAATGIEPTEA--VPPK--SPAGSSTP----------------P
MAB_3498c|M.abscessus_ATCC_199 FGPGAGATATAPAVEPG---AMPPAPPGGQNVP----------------V
***** :** . .
Mflv_4654|M.gilvum_PYR-GCK EVPVAVPPTGPTQLSAAKSAALQDVNAALDALRGAQESGDFAQYGEALQR
Mvan_1814|M.vanbaalenii_PYR-1 EVPVAVPPTGPTQLSAGKAAALQDVNAALDALQDAQRSGDFAQYGEALQR
MSMEG_1959|M.smegmatis_MC2_155 PPPSAATPTGPVQISQAKAEALQDLESALTAAQEAQRSGDFAEYGQALQR
MMAR_1371|M.marinum_M PTAVPPAPEASVTLSPAKAAAMKEIQSAIGAARDAQKKGDFAAYGAALQR
MUL_2505|M.ulcerans_Agy99 PTAVPPAPEASVTLSPARAAAMKEIQSAIGAARDAQKKGDFAAYGAALQR
MAV_4137|M.avium_104 PVAAPPVPDGSVTLSPAKAAVLQEIQAAIGAAKDAQKKGDFAGYGAALQR
Mb3215c|M.bovis_AF2122/97 VTAAPPVPVGAVTLSPAKVAALQEIQAAIGAARDAQKKGDFAAYGSALQR
Rv3193c|M.tuberculosis_H37Rv VTAAPPVPVGAVTLSPAKVAALQEIQAAIGAARDAQKKGDFAAYGSALQR
MLBr_00644|M.leprae_Br4923 IAVVPSAPDGSVALSAAKAAALQEIQAVIGAAREAQKKGDFVAYGSALQR
MAB_3498c|M.abscessus_ATCC_199 VPPVTPPPTGSAELSSAKAAALQEVQRAIGEVKEAQKSGDFARYGQALKG
. * ... :* .: .::::: .: : **..***. ** **:
Mflv_4654|M.gilvum_PYR-GCK LDDAVNKYQETD
Mvan_1814|M.vanbaalenii_PYR-1 LDDAVNKYQATN
MSMEG_1959|M.smegmatis_MC2_155 LNDAMKKYDSAK
MMAR_1371|M.marinum_M LDDAITKFNNTQ
MUL_2505|M.ulcerans_Agy99 LDDAITKFNNTQ
MAV_4137|M.avium_104 LDDAITKYNNTK
Mb3215c|M.bovis_AF2122/97 LDEAITKFNDAG
Rv3193c|M.tuberculosis_H37Rv LDEAITKFNDAG
MLBr_00644|M.leprae_Br4923 LDDAITKFNSTK
MAB_3498c|M.abscessus_ATCC_199 LDDAMTKFTQAR
*::*:.*: :