For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAPSPAQITAAPEARRLAAMLFGIGTLHFLAPKPFDSIVPAELPGSARFYTYASGVGELATGALLASPRT RRLGALAAVALFISVFPANVNMVRLWWNKPWPLRLGAIARLPLQIPMITTALKVYRHS*
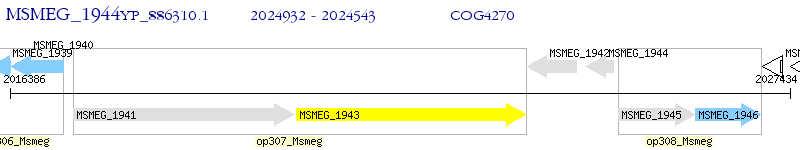
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1944 | - | - | 100% (129) | membrane protein |
| M. smegmatis MC2 155 | MSMEG_0814 | - | 2e-07 | 40.68% (59) | membrane protein |
| M. smegmatis MC2 155 | MSMEG_5840 | - | 1e-05 | 38.27% (81) | hypothetical protein MSMEG_5840 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4664 | - | 5e-41 | 59.85% (132) | hypothetical protein Mflv_4664 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0592c | - | 2e-44 | 73.45% (113) | hypothetical protein MAB_0592c |
| M. marinum M | MMAR_4907 | - | 6e-05 | 37.04% (81) | hypothetical protein MMAR_4907 |
| M. avium 104 | MAV_2126 | - | 6e-47 | 73.04% (115) | membrane protein |
| M. thermoresistible (build 8) | TH_2857 | - | 3e-06 | 44.00% (50) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_0499 | - | 3e-05 | 37.04% (81) | hypothetical protein MUL_0499 |
| M. vanbaalenii PYR-1 | Mvan_1803 | - | 2e-42 | 64.80% (125) | hypothetical protein Mvan_1803 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4664|M.gilvum_PYR-GCK ------MRQPHQHNVIMTSSPSALQRISTSSRGARMGAWLVGMGVLHFAA
Mvan_1803|M.vanbaalenii_PYR-1 ----------------MTTSQTALQQIPKSSRGARMGAWLVGMGILHFAA
MSMEG_1944|M.smegmatis_MC2_155 -----------------MTAPSPAQITAAP-EARRLAAMLFGIGTLHFLA
MAV_2126|M.avium_104 -----------------MTTRSAAARDATELAAYRVAAMLLGVGTLHFLA
MAB_0592c|M.abscessus_ATCC_199 -----------------METK------ASPWPAYRLAAMLLGVGVVHFVA
MMAR_4907|M.marinum_M MAPMITLVLGSIVARIIGQLGVAYLDSWSAAIAVGLAAMFTLTGIAHFAP
MUL_0499|M.ulcerans_Agy99 MAPMITLVLGSIVARIIGQLGVAYLDSWSAAIAVGLAAMFTLTGIAHFAP
TH_2857|M.thermoresistible__bu ----------------------------MTGNRHLLGLVVSLIGVAHFVA
:. . * ** .
Mflv_4664|M.gilvum_PYR-GCK PKP--FDTIIPEELPGSARFYTYASGVAEVGVGGLLLVPATRRIGALSAI
Mvan_1803|M.vanbaalenii_PYR-1 PKP--FDSIVPEELPGTQRFYTYASGVAEIGVGALLLAPRTRRAGALSAI
MSMEG_1944|M.smegmatis_MC2_155 PKP--FDSIVPAELPGSARFYTYASGVGELATGALLASPRTRRLGALAAV
MAV_2126|M.avium_104 PKP--FDSIIPAELPGSPRLYTYGSGVAELTVGALLVPRRTRRAAALAAA
MAB_0592c|M.abscessus_ATCC_199 PKP--FDDIVPAGLPGDPRLYTYASGVAELGTGALLLAPKTRRLGGTLAA
MMAR_4907|M.marinum_M PLRGDLVAIVPPRLP-APGLLVSITGALELLGAAGLLLPVTRVAAALCFL
MUL_0499|M.ulcerans_Agy99 PLRGDLVAIVPPRLP-APGLLVSITGALELLGAAGLLLPVTRVAAALCLL
TH_2857|M.thermoresistible__bu PRL--FDPVNRLGFPTRARTFTYVNGGLETLLGVLIAVPGTQRLSRYVS-
* : : :* . .* * . : *: .
Mflv_4664|M.gilvum_PYR-GCK ALFVGVFPGNANMVRMWFRDPTKNLAMRLGAIARLPLQVPMITQAWKIYK
Mvan_1803|M.vanbaalenii_PYR-1 ALFVGVFPGNVNMVRLWFNDPTKSLPMRVAAVARLPLQVPMIMQAWKVYR
MSMEG_1944|M.smegmatis_MC2_155 ALFISVFPANVNMVRLWWNKP---WPLRLGAIARLPLQIPMITTALKVYR
MAV_2126|M.avium_104 ILFVGVFPGNLNMVRLWWDKP---WPMRIAALARLPLQIPMITTALRIRR
MAB_0592c|M.abscessus_ATCC_199 LLFIAVFPANINMVRLWWDKP---LALKLGAIARLPMQIPMITEALKVRR
MMAR_4907|M.marinum_M ILMLAMFPANIYAARMPSPPK----SMTTRLSVRSAEELVFLAAAAIVAT
MUL_0499|M.ulcerans_Agy99 ILMLAMFPANIYAARMPSPPK----SMTTRLSVRSAEELVFLAAAAIVAT
TH_2857|M.thermoresistible__bu ACYTAYLTAAVARTQL---------STRHGRR------------------
. :.. .:: .
Mflv_4664|M.gilvum_PYR-GCK ES--
Mvan_1803|M.vanbaalenii_PYR-1 ES--
MSMEG_1944|M.smegmatis_MC2_155 HS--
MAV_2126|M.avium_104 NS--
MAB_0592c|M.abscessus_ATCC_199 TA--
MMAR_4907|M.marinum_M GGCQ
MUL_0499|M.ulcerans_Agy99 GGCQ
TH_2857|M.thermoresistible__bu ----